Paleoproteomics
- PMID: 35839101
- PMCID: PMC9412968
- DOI: 10.1021/acs.chemrev.1c00703
Paleoproteomics
Abstract
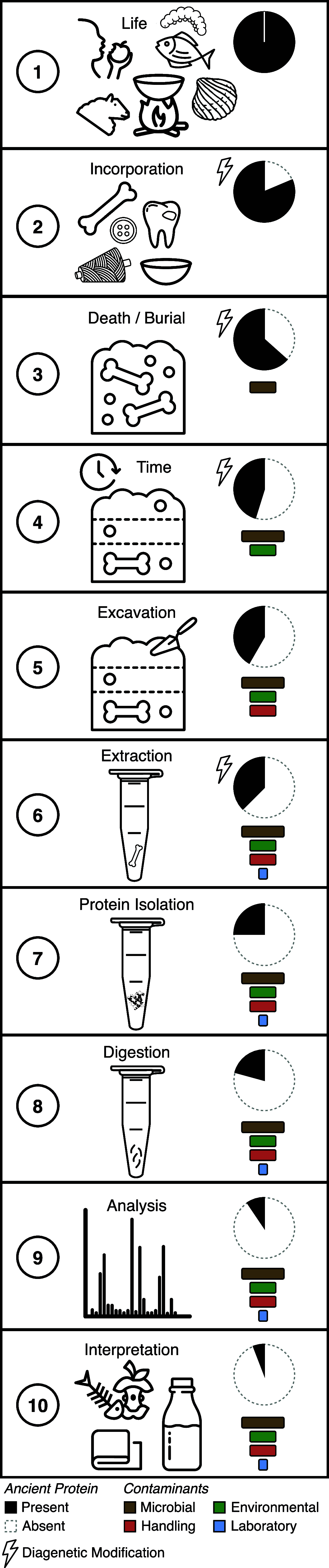
Paleoproteomics, the study of ancient proteins, is a rapidly growing field at the intersection of molecular biology, paleontology, archaeology, paleoecology, and history. Paleoproteomics research leverages the longevity and diversity of proteins to explore fundamental questions about the past. While its origins predate the characterization of DNA, it was only with the advent of soft ionization mass spectrometry that the study of ancient proteins became truly feasible. Technological gains over the past 20 years have allowed increasing opportunities to better understand preservation, degradation, and recovery of the rich bioarchive of ancient proteins found in the archaeological and paleontological records. Growing from a handful of studies in the 1990s on individual highly abundant ancient proteins, paleoproteomics today is an expanding field with diverse applications ranging from the taxonomic identification of highly fragmented bones and shells and the phylogenetic resolution of extinct species to the exploration of past cuisines from dental calculus and pottery food crusts and the characterization of past diseases. More broadly, these studies have opened new doors in understanding past human-animal interactions, the reconstruction of past environments and environmental changes, the expansion of the hominin fossil record through large scale screening of nondiagnostic bone fragments, and the phylogenetic resolution of the vertebrate fossil record. Even with these advances, much of the ancient proteomic record still remains unexplored. Here we provide an overview of the history of the field, a summary of the major methods and applications currently in use, and a critical evaluation of current challenges. We conclude by looking to the future, for which innovative solutions and emerging technology will play an important role in enabling us to access the still unexplored "dark" proteome, allowing for a fuller understanding of the role ancient proteins can play in the interpretation of the past.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Boyd W. C.; Boyd L. G. Blood Grouping Tests on 300 Mummies: With Notes on the Precipitin-Test. J. Immunol. 1937, 32, 307–319.
-
- Abelson P. H. Paleobiochemistry: Organic Constituents of Fossils. Carnegie Institution of Washington, Yearbook 1954, 53, 97–101.
-
- Ostrom P. H.; Schall M.; Gandhi H.; Shen T.-L.; Hauschka P. V.; Strahler J. R.; Gage D. A. New Strategies for Characterizing Ancient Proteins Using Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Geochim. Cosmochim. Acta 2000, 64, 1043–1050. 10.1016/S0016-7037(99)00381-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

