Explainable deep transfer learning model for disease risk prediction using high-dimensional genomic data
- PMID: 35839250
- PMCID: PMC9328574
- DOI: 10.1371/journal.pcbi.1010328
Explainable deep transfer learning model for disease risk prediction using high-dimensional genomic data
Abstract
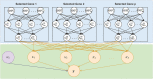
Building an accurate disease risk prediction model is an essential step in the modern quest for precision medicine. While high-dimensional genomic data provides valuable data resources for the investigations of disease risk, their huge amount of noise and complex relationships between predictors and outcomes have brought tremendous analytical challenges. Deep learning model is the state-of-the-art methods for many prediction tasks, and it is a promising framework for the analysis of genomic data. However, deep learning models generally suffer from the curse of dimensionality and the lack of biological interpretability, both of which have greatly limited their applications. In this work, we have developed a deep neural network (DNN) based prediction modeling framework. We first proposed a group-wise feature importance score for feature selection, where genes harboring genetic variants with both linear and non-linear effects are efficiently detected. We then designed an explainable transfer-learning based DNN method, which can directly incorporate information from feature selection and accurately capture complex predictive effects. The proposed DNN-framework is biologically interpretable, as it is built based on the selected predictive genes. It is also computationally efficient and can be applied to genome-wide data. Through extensive simulations and real data analyses, we have demonstrated that our proposed method can not only efficiently detect predictive features, but also accurately predict disease risk, as compared to many existing methods.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants.Mol Plant. 2023 Jan 2;16(1):279-293. doi: 10.1016/j.molp.2022.11.004. Epub 2022 Nov 10. Mol Plant. 2023. PMID: 36366781
-
Ge-SAND: an explainable deep learning-driven framework for disease risk prediction by uncovering complex genetic interactions in parallel.BMC Genomics. 2025 May 1;26(1):432. doi: 10.1186/s12864-025-11588-9. BMC Genomics. 2025. PMID: 40312319 Free PMC article.
-
A universal deep learning approach for modeling the flow of patients under different severities.Comput Methods Programs Biomed. 2018 Feb;154:191-203. doi: 10.1016/j.cmpb.2017.11.003. Epub 2017 Nov 7. Comput Methods Programs Biomed. 2018. PMID: 29249343
-
Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization.Phys Med. 2021 Mar;83:108-121. doi: 10.1016/j.ejmp.2021.03.009. Epub 2021 Mar 22. Phys Med. 2021. PMID: 33765601 Review.
-
Patient Similarity Networks for Precision Medicine.J Mol Biol. 2018 Sep 14;430(18 Pt A):2924-2938. doi: 10.1016/j.jmb.2018.05.037. Epub 2018 Jun 1. J Mol Biol. 2018. PMID: 29860027 Free PMC article. Review.
Cited by
-
Designing interpretable deep learning applications for functional genomics: a quantitative analysis.Brief Bioinform. 2024 Jul 25;25(5):bbae449. doi: 10.1093/bib/bbae449. Brief Bioinform. 2024. PMID: 39293804 Free PMC article. Review.
-
Detecting genetic interactions with visible neural networks.Commun Biol. 2025 Jun 5;8(1):874. doi: 10.1038/s42003-025-08157-x. Commun Biol. 2025. PMID: 40473911 Free PMC article.
-
Functional Neural Networks for High-Dimensional Genetic Data Analysis.IEEE/ACM Trans Comput Biol Bioinform. 2024 May-Jun;21(3):383-393. doi: 10.1109/TCBB.2024.3364614. Epub 2024 Jun 5. IEEE/ACM Trans Comput Biol Bioinform. 2024. PMID: 38507390 Free PMC article.
-
TrG2P: A transfer-learning-based tool integrating multi-trait data for accurate prediction of crop yield.Plant Commun. 2024 Jul 8;5(7):100975. doi: 10.1016/j.xplc.2024.100975. Epub 2024 May 15. Plant Commun. 2024. PMID: 38751121 Free PMC article.
-
Deep learning captures the effect of epistasis in multifactorial diseases.Front Med (Lausanne). 2025 Jan 7;11:1479717. doi: 10.3389/fmed.2024.1479717. eCollection 2024. Front Med (Lausanne). 2025. PMID: 39839630 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

