The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance
- PMID: 35840578
- PMCID: PMC9287322
- DOI: 10.1038/s41467-022-31810-6
The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance
Abstract
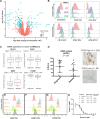
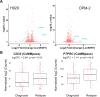
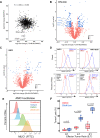
The myeloma surface proteome (surfaceome) determines tumor interaction with the microenvironment and serves as an emerging arena for therapeutic development. Here, we use glycoprotein capture proteomics to define the myeloma surfaceome at baseline, in drug resistance, and in response to acute drug treatment. We provide a scoring system for surface antigens and identify CCR10 as a promising target in this disease expressed widely on malignant plasma cells. We engineer proof-of-principle chimeric antigen receptor (CAR) T-cells targeting CCR10 using its natural ligand CCL27. In myeloma models we identify proteins that could serve as markers of resistance to bortezomib and lenalidomide, including CD53, CD10, EVI2B, and CD33. We find that acute lenalidomide treatment increases activity of MUC1-targeting CAR-T cells through antigen upregulation. Finally, we develop a miniaturized surface proteomic protocol for profiling primary plasma cell samples with low inputs. These approaches and datasets may contribute to the biological, therapeutic, and diagnostic understanding of myeloma.
© 2022. The Author(s).
Conflict of interest statement
A.P.W. is a member of the Scientific Advisory Board and holds equity stakes in Indapta Therapeutics and Protocol Intelligence, LLC. J.A.W. is on the Scientific Advisory Board and holds equity stakes in the following companies with oncology interests: Soteria Biotherapeutics, Jnana Therapeutics, Inception Therapeutics, and Inzen Therapeutics, and holds a sponsored research agreement with Bristol Myers Squibb. S.W.W. has received research funding from Janssen, GlaxoSmithKline, Bristol Myers Squibb, Genentech, and Fortis, and served as a consultant to Amgen. T.G.M. has received research funding from Sanofi, Janssen, and Amgen and has served as a consultant to GlaxoSmithKline. A.L.-G. is an employee and shareholder of Celgene/Bristol Myers Squibb. P.C. is currently an employee and shareholder of Roche/Genentech but was solely employed by UCSF during her participation on this project. A.D.P. is on the Scientific Advisory Board of Astellas Pharma, GO Therapeutics, and Stromatis Pharma, holds equity stakes in Stromatis Pharma, and received research funding from Tmunity Therapeutics. J.E. is a co-founder and holds an equity stake in Mnemo Therapeutics. The other authors declare no conflicts of interest.
Figures



Comment in
-
Unraveling the surface proteomic profile of multiple myeloma to reveal new immunotherapeutic targets and markers of drug resistance.Cell Stress. 2022 Oct 13;6(11):89-92. doi: 10.15698/cst2022.11.273. eCollection 2022 Nov. Cell Stress. 2022. PMID: 36311892 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

