Identification of multi-omics biomarkers and construction of the novel prognostic model for hepatocellular carcinoma
- PMID: 35840618
- PMCID: PMC9287549
- DOI: 10.1038/s41598-022-16341-w
Identification of multi-omics biomarkers and construction of the novel prognostic model for hepatocellular carcinoma
Abstract
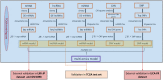
Genome changes play a crucial role in carcinogenesis, and many biomarkers can be used as effective prognostic indicators in various tumors. Although previous studies have constructed many predictive models for hepatocellular carcinoma (HCC) based on molecular signatures, the performance is unsatisfactory. Because multi-omics data can more comprehensively reflect the biological phenomenon of disease, we hope to build a more accurate predictive model by multi-omics analysis. We use the TCGA to identify crucial biomarkers and construct prognostic models through difference analysis, univariate Cox, and LASSO/stepwise Cox analysis. The performances of predictive models were evaluated and validated through survival analysis, Harrell's concordance index (C-index), receiver operating characteristic (ROC) curve, and decision curve analysis (DCA). Multiple mRNAs, lncRNAs, miRNAs, CNV genes, and SNPs were significantly associated with the prognosis of HCC. We constructed five single-omic models, and the mRNA and lncRNA models showed good performance with c-indexes over 0.70. The multi-omics model presented a robust predictive ability with a c-index over 0.77. This study identified many biomarkers that may help study underlying carcinogenesis mechanisms in HCC. In addition, we constructed multiple single-omic models and an integrated multi-omics model that may provide practical and reliable guides for prognosis assessment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma.World J Gastroenterol. 2018 Aug 14;24(30):3426-3439. doi: 10.3748/wjg.v24.i30.3426. World J Gastroenterol. 2018. PMID: 30122881 Free PMC article.
-
Identification of Potential Prognostic Long Non-Coding RNA Biomarkers for Predicting Survival in Patients with Hepatocellular Carcinoma.Cell Physiol Biochem. 2018;48(5):1854-1869. doi: 10.1159/000492507. Epub 2018 Aug 9. Cell Physiol Biochem. 2018. PMID: 30092592
-
Identification of Long Noncoding RNA Biomarkers for Hepatocellular Carcinoma Using Single-Sample Networks.Biomed Res Int. 2020 Nov 14;2020:8579651. doi: 10.1155/2020/8579651. eCollection 2020. Biomed Res Int. 2020. PMID: 33299877 Free PMC article.
-
The Role of Long Non-Coding RNA and microRNA Networks in Hepatocellular Carcinoma and Its Tumor Microenvironment.Int J Mol Sci. 2021 Sep 30;22(19):10630. doi: 10.3390/ijms221910630. Int J Mol Sci. 2021. PMID: 34638971 Free PMC article. Review.
-
The Application of Bayesian Methods in Cancer Prognosis and Prediction.Cancer Genomics Proteomics. 2022 Jan-Feb;19(1):1-11. doi: 10.21873/cgp.20298. Cancer Genomics Proteomics. 2022. PMID: 34949654 Free PMC article. Review.
Cited by
-
Unveil Intrahepatic Cholangiocarcinoma Heterogeneity through the Lens of Omics and Multi-Omics Approaches.Cancers (Basel). 2024 Aug 20;16(16):2889. doi: 10.3390/cancers16162889. Cancers (Basel). 2024. PMID: 39199659 Free PMC article. Review.
-
Deciphering KDM8 dysregulation and CpG methylation in hepatocellular carcinoma using multi-omics and machine learning.Epigenomics. 2024;16(13):961-983. doi: 10.1080/17501911.2024.2374702. Epub 2024 Jul 29. Epigenomics. 2024. PMID: 39072393 Free PMC article.
-
Machine Learning Approach and Bioinformatics Analysis Discovered Key Genomic Signatures for Hepatitis B Virus-Associated Hepatocyte Remodeling and Hepatocellular Carcinoma.Cancer Inform. 2025 Apr 16;24:11769351251333847. doi: 10.1177/11769351251333847. eCollection 2025. Cancer Inform. 2025. PMID: 40291818 Free PMC article. Review.
-
Large-Scale Machine Learning Analysis Reveals DNA Methylation and Gene Expression Response Signatures for Gemcitabine-Treated Pancreatic Cancer.Health Data Sci. 2024 Jan 8;4:0108. doi: 10.34133/hds.0108. eCollection 2024. Health Data Sci. 2024. PMID: 38486621 Free PMC article.
-
Biomarkers for Early Cancer Detection: A Landscape View of Recent Advancements, Spotlighting Pancreatic and Liver Cancers.ACS Pharmacol Transl Sci. 2024 Feb 14;7(3):586-613. doi: 10.1021/acsptsci.3c00346. eCollection 2024 Mar 8. ACS Pharmacol Transl Sci. 2024. PMID: 38481702 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

