Construction of a circular RNA-based competing endogenous RNA network to screen biomarkers related to intervertebral disc degeneration
- PMID: 35840955
- PMCID: PMC9284696
- DOI: 10.1186/s12891-022-05579-0
Construction of a circular RNA-based competing endogenous RNA network to screen biomarkers related to intervertebral disc degeneration
Abstract
Background: Intervertebral disc degeneration (IDD) is a leading cause of disability with limited treatment strategies. A better understanding of the mechanism of IDD might enable less invasive and more targeted treatments. This study aimed to identify the circular RNA (circRNA)-microRNA (miRNA)-messenger RNA (mRNA) competing endogenous RNA (ceRNA) regulatory mechanisms in IDD. METHODS : The GSE67567 microarray dataset was downloaded from the Gene Expression Omnibus database. After data preprocessing, differentially expressed circRNAs, miRNAs and mRNAs between IDD and controls were identified. A ceRNA network was constructed on the basis of the interaction between circRNAs and miRNAs, and miRNAs and mRNAs. Pathway enrichment analysis was performed on the mRNAs in the ceRNA network. Then, with 'intervertebral disc degeneration' as keywords, IDD-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were searched for in the Comparative Toxicogenomics Database.
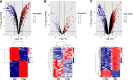
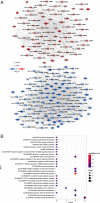
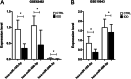
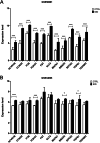
Results: A total of 105 differentially expressed circRNAs, 84 miRNAs and 967 mRNAs were identified. After analysis, 86 circRNA-miRNA, and 126 miRNA-mRNA regulatory relationship pairs were obtained to construct a ceRNA network. The mRNAs were enriched in six KEGG signalling pathways, and four were associated with IDD: the hsa04350: TGF-beta signalling pathway, hsa04068: FoxO signalling pathway, hsa05142: Chagas disease (American trypanosomiasis) and hsa04380: Osteoclast differentiation. An IDD-related ceRNA network was constructed involving four circRNAs, three miRNAs and 11 mRNAs. Auxiliary validation showed that the expression levels of miR-185-5p, miR-486-5p, ACVR1B, FOXO1, SMAD2 and TGFB1 were consistent in different databases.
Conclusions: Our study identified some circRNA-miRNA-mRNA interaction axes potentially associated with the progression of IDD, viz.: circRNA_100086-miR-509-3p-MAPK1, circRNA_000200-miR-185-5p-TGFB1, circRNA_104308-miR-185-5p-TGFB1, circRNA_400090-miR-486-5p-FOXO1 and circRNA_400090-miR-486-5p-SMAD2.
Keywords: Circular RNA; Competing endogenous RNA; Intervertebral disc degeneration; mRNA; microRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Weber KT, Jacobsen TD, Maidhof R, Virojanapa J, Overby C, Bloom O, Quraishi S, Levine M, Chahine NO. Developments in intervertebral disc disease research: pathophysiology, mechanobiology, and therapeutics. Curr Rev Musculoskelet Med. 2015;8(1):18–31. doi: 10.1007/s12178-014-9253-8. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

