The DDR-related gene signature with cell cycle checkpoint function predicts prognosis, immune activity, and chemoradiotherapy response in lung adenocarcinoma
- PMID: 35840978
- PMCID: PMC9288070
- DOI: 10.1186/s12931-022-02110-w
The DDR-related gene signature with cell cycle checkpoint function predicts prognosis, immune activity, and chemoradiotherapy response in lung adenocarcinoma
Abstract
Background: As a DNA surveillance mechanism, cell cycle checkpoint has recently been discovered to be closely associated with lung adenocarcinoma (LUAD) prognosis. It is also an essential link in the process of DNA damage repair (DDR) that confers resistance to radiotherapy. Whether genes that have both functions play a more crucial role in LUAD prognosis remains unclear.
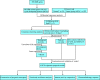
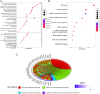
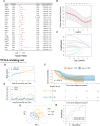
Methods: In this study, DDR-related genes with cell cycle checkpoint function (DCGs) were selected to investigate their effects on the prognosis of LUAD. The TCGA-LUAD cohort and two GEO external validation cohorts (GSE31210 and GSE42171) were performed to construct a prognosis model based on the least absolute shrinkage and selection operator (LASSO) regression. Patients were divided into high-risk and low-risk groups based on the model. Subsequently, the multivariate COX regression was used to construct a prognostic nomogram. The ssGSEA, CIBERSORT algorithm, TIMER tool, CMap database, and IC50 of chemotherapeutic agents were used to analyze immune activity and responsiveness to chemoradiotherapy.
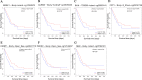
Results: 4 DCGs were selected as prognostic signatures, and patients in the high-risk group had a lower overall survival (OS). The lower infiltration levels of immune cells and the higher expression levels of immune checkpoints appeared in the high-risk group. The damage repair pathways were upregulated, and chemotherapeutic agent sensitivity was poor in the high-risk group.
Conclusions: The 4-DCGs signature prognosis model we constructed could predict the survival rate, immune activity, and chemoradiotherapy responsiveness of LUAD patients.
Keywords: Cancer prognosis; Cell cycle checkpoint; DNA damage repair; Gene signature; Lung adenocarcinoma.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Global Burden of Disease Cancer C. Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, Henrikson HJ, Lu D, Pennini A, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022;8:420–444. doi: 10.1001/jamaoncol.2021.6987. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

