Metformin combats high glucose-induced damage to the osteogenic differentiation of human periodontal ligament stem cells via inhibition of the NPR3-mediated MAPK pathway
- PMID: 35841070
- PMCID: PMC9284897
- DOI: 10.1186/s13287-022-02992-z
Metformin combats high glucose-induced damage to the osteogenic differentiation of human periodontal ligament stem cells via inhibition of the NPR3-mediated MAPK pathway
Abstract
Background: High glucose-induced damage to the osteogenic differentiation of human periodontal ligament stem cells (PDLSCs) has long been a challenge to periodontal regeneration for diabetic individuals. Metformin is an anti-hyperglycemic drug that exhibits abundant biological activities associated with cell metabolism and downstream tissue regeneration. However, how metformin combats damage to PDLSC osteogenic differentiation under high glucose and the underlying mechanisms remain unknown.
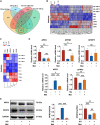
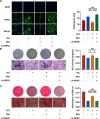
Methods: Osteogenic differentiation of PDLSCs was assessed by alkaline phosphatase (ALP) staining, ALP activity, Alizarin Red staining and quantitative assay, quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analysis. RNA-seq analysis was performed to screen target genes of metformin, and the effects of target genes were confirmed using lentivirus transfection. Western blot analysis was also used to detect the protein level of underlying signaling pathways.
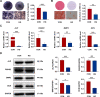
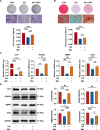
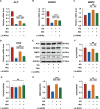
Results: We found that osteogenic differentiation of PDLSCs under high glucose was decreased, and metformin addition enhanced this capacity of differentiation. Furthermore, the results of RNA-seq analysis showed that natriuretic peptide receptor 3 (NPR3) was upregulated in PDLSCs under high glucose and downregulated after metformin addition. When the underlying pathways involved were investigated, we found that upregulation of NPR3 can compromise the metformin-enhanced PDLSC osteogenic differentiation and activate the MAPK pathway (especially the p38 MAPK and Erk1/2 pathway), and that inhibition of the NPR3-mediated p38 MAPK or Erk1/2 pathway enhanced the osteogenic differentiation of PDLSCs under high glucose.
Conclusions: The present study suggests that metformin may enhance the osteogenic differentiation of PDLSCs under high glucose via downregulation of NPR3 and inhibition of its downstream MAPK pathway. This is the first report identifying the involvement of NPR3-mediated MAPK pathway in the metformin-enhanced osteogenic differentiation, indicating that NPR3 antagonists, such as metformin, may be feasible therapeutics for periodontal tissue regeneration in diabetic individuals.
Keywords: High glucose; MAPK pathway; Metformin; NPR3; Osteogenic differentiation; Periodontal tissue regeneration.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Kinane DF. Causation and pathogenesis of periodontal disease. Periodontol. 2000;2001(25):8–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

