A structural metagenomics pipeline for examining the gut microbiome
- PMID: 35841748
- PMCID: PMC10039758
- DOI: 10.1016/j.sbi.2022.102416
A structural metagenomics pipeline for examining the gut microbiome
Abstract
Metagenomic sequencing data provide a rich resource from which to expand our understanding of differential protein functions involved in human health. Here, we outline a pipeline that combines microbial whole genome sequencing with protein structure data to yield a structural metagenomics-informed atlas of microbial enzyme families of interest. Visualizing metagenomics data through a structural lens facilitates downstream studies including targeted inhibition and probe-based proteomics to define at the molecular level how different enzyme orthologs impact in vivo function. Application of this pipeline to gut microbial enzymes like glucuronidases, TMA lyases, and bile salt hydrolases is expected to pinpoint their involvement in health and disease and may aid in the development of therapeutics that target specific enzymes within the microbiome.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement MRR is a founder of Symberix, Inc., which is developing microbiome-targeted therapeutics. MRR is also the recipient of research funding from Merck and Lilly.
Figures
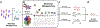
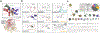


References
-
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. : An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014, 32:834–841. - PubMed
-
- Lavelle A, Sokol H: The gut microbiome in inflammatory bowel disease. In Molecular Genetics of Inflammatory Bowel Disease. Springer International Publishing; 2019:347–377.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

