Loss of immunity-related GTPase GM4951 leads to nonalcoholic fatty liver disease without obesity
- PMID: 35842425
- PMCID: PMC9288484
- DOI: 10.1038/s41467-022-31812-4
Loss of immunity-related GTPase GM4951 leads to nonalcoholic fatty liver disease without obesity
Abstract
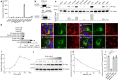
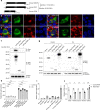
Obesity and diabetes are well known risk factors for nonalcoholic fatty liver disease (NAFLD), but the genetic factors contributing to the development of NAFLD remain poorly understood. Here we describe two semi-dominant allelic missense mutations (Oily and Carboniferous) of Predicted gene 4951 (Gm4951) identified from a forward genetic screen in mice. GM4951 deficient mice developed NAFLD on high fat diet (HFD) with no changes in body weight or glucose metabolism. Moreover, HFD caused a reduction in the level of Gm4951, which in turn promoted the development of NAFLD. Predominantly expressed in hepatocytes, GM4951 was verified as an interferon inducible GTPase. The NAFLD in Gm4951 knockout mice was associated with decreased lipid oxidation in the liver and no defect in hepatic lipid secretion. After lipid loading, hepatocyte GM4951 translocated to lipid droplets (LDs), bringing with it hydroxysteroid 17β-dehydrogenase 13 (HSD17B13), which in the absence of GM4951 did not undergo this translocation. We identified a rare non-obese mouse model of NAFLD caused by GM4951 deficiency and define a critical role for GTPase-mediated translocation in hepatic lipid metabolism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Hepatocyte-secreted FAM3D ameliorates hepatic steatosis by activating FPR1-hnRNP U-GR-SCAD pathway to enhance lipid oxidation.Metabolism. 2023 Sep;146:155661. doi: 10.1016/j.metabol.2023.155661. Epub 2023 Jul 16. Metabolism. 2023. PMID: 37454871
-
APOC3 Protein Is Not a Predisposing Factor for Fat-induced Nonalcoholic Fatty Liver Disease in Mice.J Biol Chem. 2017 Mar 3;292(9):3692-3705. doi: 10.1074/jbc.M116.765917. Epub 2017 Jan 23. J Biol Chem. 2017. PMID: 28115523 Free PMC article.
-
1,2,3,4,6 penta-O-galloyl-β-D-glucose ameliorates high-fat diet-induced nonalcoholic fatty liver disease and maintains the expression of genes involved in lipid homeostasis in mice.Biomed Pharmacother. 2020 Sep;129:110348. doi: 10.1016/j.biopha.2020.110348. Epub 2020 Jun 15. Biomed Pharmacother. 2020. PMID: 32554245
-
Rescue of Hepatic Phospholipid Remodeling Defectin iPLA2β-Null Mice Attenuates Obese but Not Non-Obese Fatty Liver.Biomolecules. 2020 Sep 17;10(9):1332. doi: 10.3390/biom10091332. Biomolecules. 2020. PMID: 32957701 Free PMC article. Review.
-
17β-hydroxysteroid dehydrogenases in the progression of nonalcoholic fatty liver disease.Pharmacol Ther. 2023 Jun;246:108428. doi: 10.1016/j.pharmthera.2023.108428. Epub 2023 Apr 26. Pharmacol Ther. 2023. PMID: 37116587 Review.
Cited by
-
A hypomorphic Mpi mutation unlocks an in vivo tool for studying global N-glycosylation deficiency.JCI Insight. 2025 Jul 22;10(14):e180752. doi: 10.1172/jci.insight.180752. eCollection 2025 Jul 22. JCI Insight. 2025. PMID: 40693465 Free PMC article.
-
Metabolic effects of CCL5 deficiency in lean and obese mice.Front Immunol. 2023 Jan 13;13:1059687. doi: 10.3389/fimmu.2022.1059687. eCollection 2022. Front Immunol. 2023. PMID: 36713454 Free PMC article.
-
Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos.Vet Sci. 2024 Aug 1;11(8):347. doi: 10.3390/vetsci11080347. Vet Sci. 2024. PMID: 39195801 Free PMC article.
-
Transcriptomics-based anti-tuberculous mechanism of traditional Chinese polyherbal preparation NiuBeiXiaoHe intermediates.Front Pharmacol. 2024 Sep 19;15:1415951. doi: 10.3389/fphar.2024.1415951. eCollection 2024. Front Pharmacol. 2024. PMID: 39364045 Free PMC article.
-
Triglycerides Mediate the Influence of Body Mass Index on Non-Alcoholic Fatty Liver Disease in a Non-Obese Chinese Population with Normal Low-Density Lipoprotein Cholesterol Levels.Obes Facts. 2024;17(2):191-200. doi: 10.1159/000536447. Epub 2024 Jan 24. Obes Facts. 2024. PMID: 38266508 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

