Metabolite, protein, and tissue dysfunction associated with COVID-19 disease severity
- PMID: 35842456
- PMCID: PMC9288092
- DOI: 10.1038/s41598-022-16396-9
Metabolite, protein, and tissue dysfunction associated with COVID-19 disease severity
Abstract
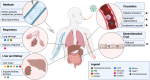
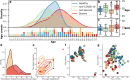
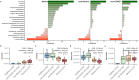
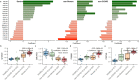
Proteins are direct products of the genome and metabolites are functional products of interactions between the host and other factors such as environment, disease state, clinical information, etc. Omics data, including proteins and metabolites, are useful in characterizing biological processes underlying COVID-19 along with patient data and clinical information, yet few methods are available to effectively analyze such diverse and unstructured data. Using an integrated approach that combines proteomics and metabolomics data, we investigated the changes in metabolites and proteins in relation to patient characteristics (e.g., age, gender, and health outcome) and clinical information (e.g., metabolic panel and complete blood count test results). We found significant enrichment of biological indicators of lung, liver, and gastrointestinal dysfunction associated with disease severity using publicly available metabolite and protein profiles. Our analyses specifically identified enriched proteins that play a critical role in responses to injury or infection within these anatomical sites, but may contribute to excessive systemic inflammation within the context of COVID-19. Furthermore, we have used this information in conjunction with machine learning algorithms to predict the health status of patients presenting symptoms of COVID-19. This work provides a roadmap for understanding the biochemical pathways and molecular mechanisms that drive disease severity, progression, and treatment of COVID-19.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
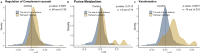
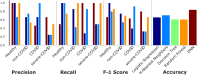
Similar articles
-
Multi-omic analysis reveals enriched pathways associated with COVID-19 and COVID-19 severity.PLoS One. 2022 Apr 25;17(4):e0267047. doi: 10.1371/journal.pone.0267047. eCollection 2022. PLoS One. 2022. PMID: 35468151 Free PMC article.
-
Leveraging metabolic modeling to identify functional metabolic alterations associated with COVID-19 disease severity.Metabolomics. 2022 Jul 11;18(7):51. doi: 10.1007/s11306-022-01904-9. Metabolomics. 2022. PMID: 35819731 Free PMC article.
-
COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19.Int J Mol Sci. 2022 Feb 22;23(5):2414. doi: 10.3390/ijms23052414. Int J Mol Sci. 2022. PMID: 35269564 Free PMC article. Review.
-
Exploring the "gene-protein-metabolite" network of coronary heart disease with phlegm and blood stasis syndrome by integrated multi-omics strategy.Front Pharmacol. 2022 Nov 29;13:1022627. doi: 10.3389/fphar.2022.1022627. eCollection 2022. Front Pharmacol. 2022. PMID: 36523490 Free PMC article.
-
Proteomics and metabolomics characterizing the pathophysiology of adaptive reactions to the metabolic challenges during the transition from late pregnancy to early lactation in dairy cows.J Proteomics. 2018 Apr 30;178:92-106. doi: 10.1016/j.jprot.2017.10.010. Epub 2017 Oct 18. J Proteomics. 2018. PMID: 29055723 Review.
Cited by
-
Crosstalk between COVID-19 Infection and Kidney Diseases: A Review on the Metabolomic Approaches.Vaccines (Basel). 2023 Feb 20;11(2):489. doi: 10.3390/vaccines11020489. Vaccines (Basel). 2023. PMID: 36851366 Free PMC article. Review.
-
Hospital antimicrobial stewardship: profiling the oral microbiome after exposure to COVID-19 and antibiotics.Front Microbiol. 2024 Feb 27;15:1346762. doi: 10.3389/fmicb.2024.1346762. eCollection 2024. Front Microbiol. 2024. PMID: 38476940 Free PMC article.
-
Gut microbiome shifts in adolescents after sleeve gastrectomy with increased oral-associated taxa and pro-inflammatory potential.Gut Microbes. 2025 Dec;17(1):2467833. doi: 10.1080/19490976.2025.2467833. Epub 2025 Feb 19. Gut Microbes. 2025. PMID: 39971742 Free PMC article.
-
Mapping Thrombosis Serum Markers by 1H-NMR Allied with Machine Learning Tools.Molecules. 2024 Dec 13;29(24):5895. doi: 10.3390/molecules29245895. Molecules. 2024. PMID: 39769984 Free PMC article.
-
A systematic review on the selection of reference genes for gene expression studies in rodents: are the classics the best choice?Mol Biol Rep. 2024 Sep 26;51(1):1017. doi: 10.1007/s11033-024-09950-3. Mol Biol Rep. 2024. PMID: 39327364
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

