Comparative transcriptomic analysis of maize ear heterosis during the inflorescence meristem differentiation stage
- PMID: 35843937
- PMCID: PMC9290290
- DOI: 10.1186/s12870-022-03695-6
Comparative transcriptomic analysis of maize ear heterosis during the inflorescence meristem differentiation stage
Abstract
Background: Heterosis is widely used in many crops and is important for global food safety, and maize is one of the most successful crops to take advantage of heterosis. Gene expression patterns control the development of the maize ear, but the mechanisms by which heterosis affects transcriptional-level control are not fully understood.
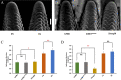
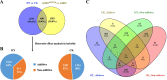
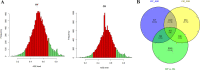
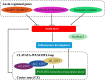
Results: In this study, we sampled ear inflorescence meristems (IMs) from the single-segment substitution maize (Zea mays) line lx9801hlEW2b, which contains the heterotic locus hlEW2b associated with ear width, as well as the receptor parent lx9801, the test parent Zheng58, and their corresponding hybrids Zheng58 × lx9801hlEW2b (HY) and Zheng58 × lx9801 (CK). After RNA sequencing and transcriptomic analysis, 2531 unique differentially expressed genes (DEGs) were identified between the two hybrids (HY vs. CK). Our results showed that approximately 64% and 48% of DEGs exhibited additive expression in HY and CK, whereas the other genes displayed a non-additive expression pattern. The DEGs were significantly enriched in GO functional categories of multiple metabolic processes, plant organ morphogenesis, and hormone regulation. These essential processes are potentially associated with heterosis performance during the maize ear developmental stage. In particular, 125 and 100 DEGs from hybrids with allele-specific expression (ASE) were specifically identified in HY and CK, respectively. Comparison between the two hybrids suggested that ASE genes were involved in different development-related processes that may lead to the hybrid vigor phenotype during maize ear development. In addition, several critical genes involved in auxin metabolism and IM development were differentially expressed between the hybrids and showed various expression patterns (additive, non-additive, and ASE). Changes in the expression levels of these genes may lead to differences in auxin homeostasis in the IM, affecting the transcription of core genes such as WUS that control IM development.
Conclusions: Our research suggests that additive, non-additive, and allele-specific expression patterns may fine-tune the expression of crucial DEGs that modulate carbohydrate and protein metabolic processes, nitrogen assimilation, and auxin metabolism to optimal levels, and these transcriptional changes may play important roles in maize ear heterosis. The results provide new information that increases our understanding of the relationship between transcriptional variation and heterosis during maize ear development, which may be helpful for clarifying the genetic and molecular mechanisms of heterosis.
Keywords: Additive and non-additive gene expression; Allele-specific expression; Heterosis; Inflorescence meristem; Maize (Zea mays L.); Transcriptomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Sokolov BP. Heterosis and maize breeding. Byul Vses n i in-ta kukuruzy. 1970:5–12.
-
- Kul'Pinova EP. Breeding sorghum for heterosis. Breeding sorghum for heterosis. 1966.
-
- Sernyk JL, Stefansson BR. Heterosis in summer rape (Brassica napus L.) Canadian Journal of Plant Science. 1983;63(2):407–413. doi: 10.4141/cjps83-046. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

