Mechanistic Models of Protein Aggregation Across Length-Scales and Time-Scales: From the Test Tube to Neurodegenerative Disease
- PMID: 35844223
- PMCID: PMC9281552
- DOI: 10.3389/fnins.2022.909861
Mechanistic Models of Protein Aggregation Across Length-Scales and Time-Scales: From the Test Tube to Neurodegenerative Disease
Abstract
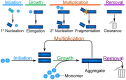
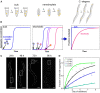
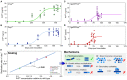
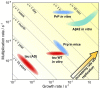
Through advances in the past decades, the central role of aberrant protein aggregation has been established in many neurodegenerative diseases. Crucially, however, the molecular mechanisms that underlie aggregate proliferation in the brains of affected individuals are still only poorly understood. Under controlled in vitro conditions, significant progress has been made in elucidating the molecular mechanisms that take place during the assembly of purified protein molecules, through advances in both experimental methods and the theories used to analyse the resulting data. The determination of the aggregation mechanism for a variety of proteins revealed the importance of intermediate oligomeric species and of the interactions with promotors and inhibitors. Such mechanistic insights, if they can be achieved in a disease-relevant system, provide invaluable information to guide the design of potential cures to these devastating disorders. However, as experimental systems approach the situation present in real disease, their complexity increases substantially. Timescales increase from hours an aggregation reaction takes in vitro, to decades over which the process takes place in disease, and length-scales increase to the dimension of a human brain. Thus, molecular level mechanistic studies, like those that successfully determined mechanisms in vitro, have only been applied in a handful of living systems to date. If their application can be extended to further systems, including patient data, they promise powerful new insights. Here we present a review of the existing strategies to gain mechanistic insights into the molecular steps driving protein aggregation and discuss the obstacles and potential paths to achieving their application in disease. First, we review the experimental approaches and analysis techniques that are used to establish the aggregation mechanisms in vitro and the insights that have been gained from them. We then discuss how these approaches must be modified and adapted to be applicable in vivo and review the existing works that have successfully applied mechanistic analysis of protein aggregation in living systems. Finally, we present a broad mechanistic classification of in vivo systems and discuss what will be required to further our understanding of aggregate formation in living systems.
Keywords: amyloid; chemical kinetics; in vivo models; mechanistic models; neurodegenerative disease; protein aggregation.
Copyright © 2022 Meisl, Knowles and Klenerman.
Conflict of interest statement
GM is a data scientist and TK is a co-founder at Wren Therapeutics. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The thermodynamics of neurodegenerative disease.Biophys Rev (Melville). 2024 Mar 20;5(1):011303. doi: 10.1063/5.0180899. eCollection 2024 Mar. Biophys Rev (Melville). 2024. PMID: 38525484 Free PMC article. Review.
-
Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange.Acc Chem Res. 2010 Aug 17;43(8):1072-9. doi: 10.1021/ar9002784. Acc Chem Res. 2010. PMID: 20557067
-
Kinetic models reveal the interplay of protein production and aggregation.Chem Sci. 2024 May 10;15(22):8430-8442. doi: 10.1039/d4sc00088a. eCollection 2024 Jun 5. Chem Sci. 2024. PMID: 38846392 Free PMC article.
-
Chemical strategies for controlling protein folding and elucidating the molecular mechanisms of amyloid formation and toxicity.J Mol Biol. 2012 Aug 10;421(2-3):204-36. doi: 10.1016/j.jmb.2012.01.051. Epub 2012 Feb 14. J Mol Biol. 2012. PMID: 22342932 Review.
-
Probing protein aggregation through spectroscopic insights and multimodal approaches: A comprehensive review for counteracting neurodegenerative disorders.Heliyon. 2024 Mar 18;10(7):e27949. doi: 10.1016/j.heliyon.2024.e27949. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38689955 Free PMC article. Review.
Cited by
-
A mechanistic model of pure and lipidic α-synuclein aggregation for advancing Parkinson's therapies.Commun Chem. 2025 Jun 14;8(1):186. doi: 10.1038/s42004-025-01558-3. Commun Chem. 2025. PMID: 40517155 Free PMC article.
-
Multiscale Simulations of Self-Assembling Peptides: Surface and Core Hydrophobicity Determine Fibril Stability and Amyloid Aggregation.Biomacromolecules. 2024 May 13;25(5):3063-3075. doi: 10.1021/acs.biomac.4c00151. Epub 2024 Apr 23. Biomacromolecules. 2024. PMID: 38652055 Free PMC article.
-
Molecular mechanism of α-synuclein aggregation on lipid membranes revealed.Chem Sci. 2024 Apr 22;15(19):7229-7242. doi: 10.1039/d3sc05661a. eCollection 2024 May 15. Chem Sci. 2024. PMID: 38756798 Free PMC article.
-
The thermodynamics of neurodegenerative disease.Biophys Rev (Melville). 2024 Mar 20;5(1):011303. doi: 10.1063/5.0180899. eCollection 2024 Mar. Biophys Rev (Melville). 2024. PMID: 38525484 Free PMC article. Review.
-
Decoupling Phase Separation and Fibrillization Preserves Activity of Biomolecular Condensates.bioRxiv [Preprint]. 2025 Mar 19:2025.03.18.643977. doi: 10.1101/2025.03.18.643977. bioRxiv. 2025. PMID: 40166274 Free PMC article. Preprint.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

