Multiplexed Visualization Method to Explore Complete Targeting Regulatory Relationships Among Circadian Genes for Insomnia Treatment
- PMID: 35844237
- PMCID: PMC9285005
- DOI: 10.3389/fnins.2022.877802
Multiplexed Visualization Method to Explore Complete Targeting Regulatory Relationships Among Circadian Genes for Insomnia Treatment
Abstract
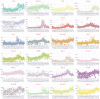
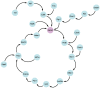
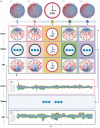
Understanding the complete map of melatonin synthesis, the information transfer network among circadian genes in pineal gland, promises to resolve outstanding issues in endocrine systems and improve the clinical diagnosis and treatment level of insomnia, immune disease and hysterical depression. Currently, some landmark studies have revealed some genes that regulate circadian rhythm associated with melatonin synthesis. However, these studies don't give a complete map of melatonin synthesis, as transfer information among circadian genes in pineal gland is lost. New biotechnology, integrates dynamic sequential omics and multiplexed imaging method, has been used to visualize the complete process of melatonin synthesis. It is found that there are two extremely significant information transfer processes involved in melatonin synthesis. In the first stage, as the light intensity decreased, melatonin synthesis mechanism has started, which is embodied in circadian genes, Rel, Polr2A, Mafk, and Srbf1 become active. In the second stage, circadian genes Hif1a, Bach1, Clock, E2f6, and Per2 are regulated simultaneously by four genes, Rel, Polr2A, Mafk, and Srbf1 and contribute genetic information to Aanat. The expeditious growth in this technique offer reference for an overall understanding of gene-to-gene regulatory relationship among circadian genes in pineal gland. In the study, dynamic sequential omics and the analysis process well provide the current state and future perspectives to better diagnose and cure diseases associated with melatonin synthesis disorder.
Keywords: circadian genes; dynamic sequential omics; insomnia; melatonin synthesis; transfer information; visualization method.
Copyright © 2022 Li, Liu, Wang, Zuo, Wang, Ju, Wang, Xing, Ling, Liu, Zhang, Zhou, Yin, Cao and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Castro J. C., Valdes I., Gonzalez-Garcia L. N., Danies G., Canas S., Winck F. V., et al. (2019). Gene regulatory networks on transfer entropy (GRNTE): a novel approach to reconstruct gene regulatory interactions applied to a case study for the plant pathogen phytophthora infestans. Theor. Biol. Med. Modell. 16, 7. 10.1186/s12976-019-0103-7 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

