Metagenomic analysis of bacterial communities of Wadi Namar Lake, Riyadh, Saudi Arabia
- PMID: 35844383
- PMCID: PMC9280250
- DOI: 10.1016/j.sjbs.2022.03.001
Metagenomic analysis of bacterial communities of Wadi Namar Lake, Riyadh, Saudi Arabia
Abstract
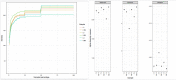
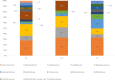
Wadi Namar lake is a new touristic attraction area in the south of Riyadh. Human activities around the lake may lead to changes in water quality with subsequent changes in microenvironment components including microbial diversity. The current study was designed to assess possible changes in bacterial communities of the water at Wadi Namar Lake. Therefore, water samples were collected from three different locations along the lake: L1 (no human activities, no plants), L2 (no human activity, some plants) and L3 (human activities, municipal wastes and some plants). The total DNA of the samples was extracted and subjected to 16S rDNA sequencing and metagenomic analysis; water pH, electrical conductivity (EC), total dissolved solids (TDS) as well as the concentration of Na+1, K+1, Cl-1 and total N were analysed. Metagenomic analysis showed variations in relative abundance of 17 phyla, 31 families, 43 genera and 19 species of bacteria between the locations. Proteobacteria was the most abundant phylum in all locations; however, its highest abundance was in L1. Planctomycete phylum was highly abundant in L1 and L3, while its abundance in L2 was low. The phyla Acidobacteria, Candidatus Saccharibacteria, Nitrospirae and Chloroflexi were associated with high TDS, EC, K+1 and Cl-1 concentrations in L3; various human activities around this location had possibly affected microbial diversity. Current study results help in recognising the structure of bacterial communities at Wadi Namar Lake in relation to their surroundings for planning to environment protection and future restoration of affected ecosystems.
Keywords: 16S rRNA gene; Bacterial abundance; Environment; Metagenomics; Water microbiome.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Metagenomic Analysis of Sediment Bacterial Diversity and Composition in Natural Lakes and Artificial Waterpoints of Tabuk Region in King Salman Bin Abdulaziz Royal Natural Reserve, Saudi Arabia.Life (Basel). 2024 Nov 1;14(11):1411. doi: 10.3390/life14111411. Life (Basel). 2024. PMID: 39598209 Free PMC article.
-
Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi An Natrun, Egypt.Microb Ecol. 2007 Nov;54(4):598-617. doi: 10.1007/s00248-006-9193-y. Epub 2007 Apr 21. Microb Ecol. 2007. PMID: 17450395
-
Diversity and Composition of Bacterial Community in Soils and Lake Sediments from an Arctic Lake Area.Front Microbiol. 2016 Jul 28;7:1170. doi: 10.3389/fmicb.2016.01170. eCollection 2016. Front Microbiol. 2016. PMID: 27516761 Free PMC article.
-
Evaluation of microbial diversity of three recreational water bodies using 16S rRNA metagenomic approach.Sci Total Environ. 2021 Jun 1;771:144773. doi: 10.1016/j.scitotenv.2020.144773. Epub 2021 Jan 26. Sci Total Environ. 2021. PMID: 33548724
-
Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam.Int J Environ Res Public Health. 2016 Jun 21;13(6):617. doi: 10.3390/ijerph13060617. Int J Environ Res Public Health. 2016. PMID: 27338434 Free PMC article.
Cited by
-
Metagenomics Analysis of the Microbial Consortium in Samples from Lake Xochimilco, a World Cultural Heritage Site.Microorganisms. 2025 Apr 7;13(4):835. doi: 10.3390/microorganisms13040835. Microorganisms. 2025. PMID: 40284671 Free PMC article.
References
-
- Ahmad T., Singh R.S., Gupta G., Sharma A., Kaur B. Adva. Enzy. Tech. Elsevier; 2019. Metagenomics in the search for industrial enzymes; pp. 419–451.
LinkOut - more resources
Full Text Sources

