Epigenetic Aspects and Prospects in Autoimmune Hepatitis
- PMID: 35844554
- PMCID: PMC9281562
- DOI: 10.3389/fimmu.2022.921765
Epigenetic Aspects and Prospects in Autoimmune Hepatitis
Abstract
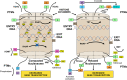
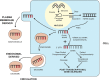
The observed risk of autoimmune hepatitis exceeds its genetic risk, and epigenetic factors that alter gene expression without changing nucleotide sequence may help explain the disparity. Key objectives of this review are to describe the epigenetic modifications that affect gene expression, discuss how they can affect autoimmune hepatitis, and indicate prospects for improved management. Multiple hypo-methylated genes have been described in the CD4+ and CD19+ T lymphocytes of patients with autoimmune hepatitis, and the circulating micro-ribonucleic acids, miR-21 and miR-122, have correlated with laboratory and histological features of liver inflammation. Both epigenetic agents have also correlated inversely with the stage of liver fibrosis. The reduced hepatic concentration of miR-122 in cirrhosis suggests that its deficiency may de-repress the pro-fibrotic prolyl-4-hydroxylase subunit alpha-1 gene. Conversely, miR-155 is over-expressed in the liver tissue of patients with autoimmune hepatitis, and it may signify active immune-mediated liver injury. Different epigenetic findings have been described in diverse autoimmune and non-autoimmune liver diseases, and these changes may have disease-specificity. They may also be responses to environmental cues or heritable adaptations that distinguish the diseases. Advances in epigenetic editing and methods for blocking micro-ribonucleic acids have improved opportunities to prove causality and develop site-specific, therapeutic interventions. In conclusion, the role of epigenetics in affecting the risk, clinical phenotype, and outcome of autoimmune hepatitis is under-evaluated. Full definition of the epigenome of autoimmune hepatitis promises to enhance understanding of pathogenic mechanisms and satisfy the unmet clinical need to improve therapy for refractory disease.
Keywords: autoimmune; chromatin modifications; epigenome; hepatitis; micro-ribonucleic acids; treatment.
Copyright © 2022 Czaja.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor NK declared a past co-authorship with the author.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

