A Path-Based Analysis of Infected Cell Line and COVID-19 Patient Transcriptome Reveals Novel Potential Targets and Drugs Against SARS-CoV-2
- PMID: 35844595
- PMCID: PMC9284228
- DOI: 10.3389/fimmu.2022.918817
A Path-Based Analysis of Infected Cell Line and COVID-19 Patient Transcriptome Reveals Novel Potential Targets and Drugs Against SARS-CoV-2
Abstract
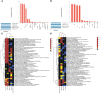
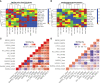
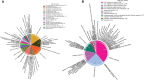
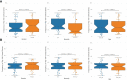
Most transcriptomic studies of SARS-CoV-2 infection have focused on differentially expressed genes, which do not necessarily reveal the genes mediating the transcriptomic changes. In contrast, exploiting curated biological network, our PathExt tool identifies central genes from the differentially active paths mediating global transcriptomic response. Here we apply PathExt to multiple cell line infection models of SARS-CoV-2 and other viruses, as well as to COVID-19 patient-derived PBMCs. The central genes mediating SARS-CoV-2 response in cell lines were uniquely enriched for ATP metabolic process, G1/S transition, leukocyte activation and migration. In contrast, PBMC response reveals dysregulated cell-cycle processes. In PBMC, the most frequently central genes are associated with COVID-19 severity. Importantly, relative to differential genes, PathExt-identified genes show greater concordance with several benchmark anti-COVID-19 target gene sets. We propose six novel anti-SARS-CoV-2 targets ADCY2, ADSL, OCRL, TIAM1, PBK, and BUB1, and potential drugs targeting these genes, such as Bemcentinib, Phthalocyanine, and Conivaptan.
Keywords: DEGs (Differentially Expressed Genes); PBMCs (Peripheral Blood Mononuclear Cells); SARS-C0V-2; cell lines; network analysis; transcriptome.
Copyright © 2022 Agrawal, Sambaturu, Olgun and Hannenhalli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Update of
-
A path-based analysis of infected cell line and COVID-19 patient transcriptome reveals novel potential targets and drugs against SARS-CoV-2.Res Sq [Preprint]. 2022 Mar 21:rs.3.rs-1474136. doi: 10.21203/rs.3.rs-1474136/v1. Res Sq. 2022. Update in: Front Immunol. 2022 Jul 01;13:918817. doi: 10.3389/fimmu.2022.918817. PMID: 35434729 Free PMC article. Updated. Preprint.
Similar articles
-
A path-based analysis of infected cell line and COVID-19 patient transcriptome reveals novel potential targets and drugs against SARS-CoV-2.Res Sq [Preprint]. 2022 Mar 21:rs.3.rs-1474136. doi: 10.21203/rs.3.rs-1474136/v1. Res Sq. 2022. Update in: Front Immunol. 2022 Jul 01;13:918817. doi: 10.3389/fimmu.2022.918817. PMID: 35434729 Free PMC article. Updated. Preprint.
-
Comparative Transcriptomic Analyses of Peripheral Blood Mononuclear Cells of COVID-19 Patients without Pneumonia and with Severe Pneumonia in the First Year of Follow-Up.Viruses. 2024 Jul 28;16(8):1211. doi: 10.3390/v16081211. Viruses. 2024. PMID: 39205185 Free PMC article.
-
Human Endogenous Retrovirus (HERV) Transcriptome Is Dynamically Modulated during SARS-CoV-2 Infection and Allows Discrimination of COVID-19 Clinical Stages.Microbiol Spectr. 2023 Feb 14;11(1):e0251622. doi: 10.1128/spectrum.02516-22. Epub 2023 Jan 5. Microbiol Spectr. 2023. PMID: 36602345 Free PMC article.
-
Detection of Antibodies Against the SARS-CoV-2 Spike Protein and Analysis of the Peripheral Blood Mononuclear Cell Transcriptomic Profile, 15 Years After Recovery From SARS.Front Cell Infect Microbiol. 2021 Nov 18;11:768993. doi: 10.3389/fcimb.2021.768993. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34869070 Free PMC article.
-
Gene Network Analysis of the Transcriptome Impact of SARS-CoV-2 Interacting MicroRNAs in COVID-19 Disease.Int J Mol Sci. 2022 Aug 17;23(16):9239. doi: 10.3390/ijms23169239. Int J Mol Sci. 2022. PMID: 36012503 Free PMC article. Review.
Cited by
-
Identification of critical genes and molecular pathways in COVID-19 myocarditis and constructing gene regulatory networks by bioinformatic analysis.PLoS One. 2022 Jun 24;17(6):e0269386. doi: 10.1371/journal.pone.0269386. eCollection 2022. PLoS One. 2022. PMID: 35749386 Free PMC article.
-
Identification and evaluation of candidate COVID-19 critical genes and medicinal drugs related to plasma cells.BMC Infect Dis. 2024 Oct 3;24(1):1099. doi: 10.1186/s12879-024-10000-3. BMC Infect Dis. 2024. PMID: 39363208 Free PMC article.
-
SARS-CoV-2 nucleocapsid protein promotes self-deacetylation by inducing HDAC6 to facilitate viral replication.Virol J. 2024 Aug 12;21(1):186. doi: 10.1186/s12985-024-02460-5. Virol J. 2024. PMID: 39135075 Free PMC article.
-
Network-based approach identifies key genes associated with tumor heterogeneity in HPV positive and negative head and neck cancer patients.Sci Rep. 2025 Aug 7;15(1):28864. doi: 10.1038/s41598-025-13604-0. Sci Rep. 2025. PMID: 40775425 Free PMC article.
-
Exploration of the potential common pathogenic mechanisms in COVID-19 and silicosis by using bioinformatics and system biology.Funct Integr Genomics. 2023 Jun 6;23(3):199. doi: 10.1007/s10142-023-01092-2. Funct Integr Genomics. 2023. PMID: 37278873 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

