A Combination of Membrane Filtration and Raman-Active DNA Ligand Greatly Enhances Sensitivity of SERS-Based Aptasensors for Influenza A Virus
- PMID: 35844641
- PMCID: PMC9279936
- DOI: 10.3389/fchem.2022.937180
A Combination of Membrane Filtration and Raman-Active DNA Ligand Greatly Enhances Sensitivity of SERS-Based Aptasensors for Influenza A Virus
Abstract
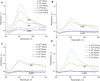
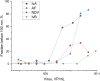
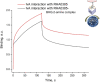
Biosensors combining the ultrahigh sensitivity of surface-enhanced Raman scattering (SERS) and the specificity of nucleic acid aptamers have recently drawn attention in the detection of respiratory viruses. The most sensitive SERS-based aptasensors allow determining as low as 104 virus particles per mL that is 100-fold lower than any antibody-based lateral flow tests but 10-100-times higher than a routine polymerase chain reaction with reversed transcription (RT-PCR). Sensitivity of RT-PCR has not been achieved in SERS-based aptasensors despite the usage of sophisticated SERS-active substrates. Here, we proposed a novel design of a SERS-based aptasensor with the limit of detection of just 103 particles per ml of the influenza A virus that approaches closely to RT-PCR sensitivity. The sensor utilizes silver nanoparticles with the simplest preparation instead of sophisticated SERS-active surfaces. The analytical signal is provided by a unique Raman-active dye that competes with the virus for the binding to the G-quadruplex core of the aptamer. The aptasensor functions even with aliquots of the biological fluids due to separation of the off-target molecules by pre-filtration through a polymeric membrane. The aptasensor detects influenza viruses in the range of 1·103-5·1010 virus particles per ml.
Keywords: SERS; aptamer; colloidal nanoparticle; influenza; membrane; sensor; virus.
Copyright © 2022 Zhdanov, Nyhrikova, Meshcheryakova, Kristavchuk, Akhmetova, Andreev, Rudakova, Gambaryan, Yaminsky, Aralov, Kukushkin and Zavyalova.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Apel P. Y. (1995). Heavy Particle Tracks in Polymers and Polymeric Track Membranes. Radiat. Meas. 25, 667–674. 10.1016/1350-4487(95)00219-5 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

