Interaction between BEND5 and RBPJ suppresses breast cancer growth and metastasis via inhibiting Notch signaling
- PMID: 35844785
- PMCID: PMC9274485
- DOI: 10.7150/ijbs.70866
Interaction between BEND5 and RBPJ suppresses breast cancer growth and metastasis via inhibiting Notch signaling
Abstract
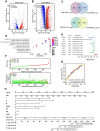
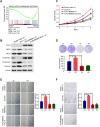
High frequent metastasis is the major cause of breast cancer (BC) mortality among women. However, the molecular mechanisms underlying BC metastasis remain largely unknown. Here, we identified six hub BC metastasis driver genes (BEND5, HSD11B1, NEDD9, SAA2, SH2D2A and TNFSF4) through bioinformatics analysis, among which BEND5 is the most significant gene. Low BEND5 expression predicted advanced stage and shorter overall survival in BC patients. Functional experiments showed that BEND5 could suppress BC growth and metastasis in vitro and in vivo. Mechanistically, BEND5 inhibits Notch signaling via directly interacting with transcription factor RBPJ/CSL. BEN domain of BEND5 interacts with the N-terminal domain (NTD) domain of RBPJ, thus preventing mastermind like transcriptional coactivator (MAML) from forming a transcription activation complex with RBPJ. Our study provides a novel insight into regulatory mechanisms underlying Notch signaling and suggests that BEND5 may become a promising target for BC therapy.
Keywords: BEND5; Breast cancer; Notch signaling; bioinformatics; metastasis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




Similar articles
-
The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.Nucleic Acids Res. 2022 Dec 9;50(22):13083-13099. doi: 10.1093/nar/gkac1137. Nucleic Acids Res. 2022. PMID: 36477367 Free PMC article.
-
RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1.EMBO J. 2017 Nov 2;36(21):3232-3249. doi: 10.15252/embj.201796525. Epub 2017 Oct 13. EMBO J. 2017. PMID: 29030483 Free PMC article.
-
Down regulation of CSL activity inhibits cell proliferation in prostate and breast cancer cells.J Cell Biochem. 2011 Sep;112(9):2340-51. doi: 10.1002/jcb.23157. J Cell Biochem. 2011. PMID: 21520243
-
CSL-Associated Corepressor and Coactivator Complexes.Adv Exp Med Biol. 2018;1066:279-295. doi: 10.1007/978-3-319-89512-3_14. Adv Exp Med Biol. 2018. PMID: 30030832 Review.
-
Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response.Adv Exp Med Biol. 2021;1287:9-30. doi: 10.1007/978-3-030-55031-8_2. Adv Exp Med Biol. 2021. PMID: 33034023 Review.
Cited by
-
The correlation between cancer stem cells and epithelial-mesenchymal transition: molecular mechanisms and significance in cancer theragnosis.Front Immunol. 2024 Sep 30;15:1417201. doi: 10.3389/fimmu.2024.1417201. eCollection 2024. Front Immunol. 2024. PMID: 39403386 Free PMC article. Review.
-
Investigation of the function of the novel tumor marker BEND5 in lung adenocarcinoma based on data mining and in vitro analysis.J Thorac Dis. 2023 Apr 28;15(4):1749-1769. doi: 10.21037/jtd-23-314. Epub 2023 Apr 10. J Thorac Dis. 2023. PMID: 37197545 Free PMC article.
-
Heterogeneity analysis and prognostic model construction of HPV negative oral squamous cell carcinoma T cells using ScRNA-seq and bulk-RNA analysis.Funct Integr Genomics. 2025 Jan 23;25(1):25. doi: 10.1007/s10142-024-01525-6. Funct Integr Genomics. 2025. PMID: 39849233 Free PMC article.
-
Epigenetic silencing of BEND4, a novel DNA damage repair gene, is a synthetic lethal marker for ATM inhibitor in pancreatic cancer.Front Med. 2024 Aug;18(4):721-734. doi: 10.1007/s11684-023-1053-3. Epub 2024 Jun 27. Front Med. 2024. PMID: 38926248
-
Clofazimine enhances anti-PD-1 immunotherapy in glioblastoma by inhibiting Wnt6 signaling and modulating the tumor immune microenvironment.Cancer Immunol Immunother. 2025 Mar 7;74(4):137. doi: 10.1007/s00262-025-03994-5. Cancer Immunol Immunother. 2025. PMID: 40053076 Free PMC article.
References
-
- DeSantis C, Ma J, Gaudet M. et al. Breast cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69:438–51. - PubMed
-
- Sung H, Ferlay J, Siegel R. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209–49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

