Identification of Prognostic Markers for Head and NeckSquamous Cell Carcinoma Based on Glycolysis-Related Genes
- PMID: 35845594
- PMCID: PMC9283050
- DOI: 10.1155/2022/2762595
Identification of Prognostic Markers for Head and NeckSquamous Cell Carcinoma Based on Glycolysis-Related Genes
Abstract
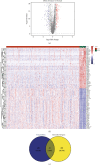
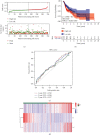
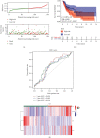
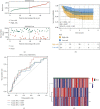
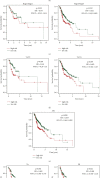
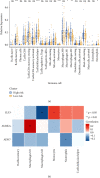
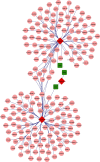
Head and neck squamous cell carcinomas (HNSCCs) comprise a heterogeneous group of tumors. Many patients respond differently to treatment and prognosis due to molecular heterogeneity. There is an urgent need to identify novel biomarkers to predict the prognosis of patients with HNSCC. Glycolysis has an important influence on the progress of HNSCC. Therefore, we investigated the prognostic significance of glycolysis-related genes in HNSCC. Our results showed that ELF3, AURKA, and ADH7 of 20 glycolysis-related DEGs were significantly related to survival and were used to construct the risk signature. The risk score showed high accuracy in distinguishing the overall survival (OS) of HNSCC. The Kaplan-Meier curves demonstrated that the risk score was associated with an unfavorable prognosis in patients with female sex, male sex, grade 3, T1/2 stage, N+ stage, N2 stage, M0 stage, and clinical stage III/IV. Independent prognostic analysis showed that clinical stage and risk score were strongly associated with OS. Moreover, the risk score had higher accuracy in predicting 1-, 3-, and 5-year survival. AURKA and ADH7 were only significantly related to M1 macrophages and neutrophils, respectively, while ELF3 was significantly correlated with M2 macrophages and monocytes (all p < 0.05).The ceRNA network demonstrated that miR-335-5p and miR-9-5p may play core roles in the regulation of these three genes in HNSCC. The risk score constructed based on three glycolysis-related genes showed high accuracy in predicting the prognosis and clinicopathological characteristics of HNSCC.
Copyright © 2022 Xiaobin Ren et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Six Glycolysis-Related Genes as Prognostic Risk Markers Can Predict the Prognosis of Patients with Head and Neck Squamous Cell Carcinoma.Biomed Res Int. 2021 Feb 10;2021:8824195. doi: 10.1155/2021/8824195. eCollection 2021. Biomed Res Int. 2021. PMID: 33628816 Free PMC article.
-
Transcriptome analysis reveals the prognostic and immune infiltration characteristics of glycolysis and hypoxia in head and neck squamous cell carcinoma.BMC Cancer. 2022 Mar 31;22(1):352. doi: 10.1186/s12885-022-09449-9. BMC Cancer. 2022. PMID: 35361159 Free PMC article.
-
A Novel Inflammatory Response-Related Gene Signature Improves High-Risk Survival Prediction in Patients With Head and Neck Squamous Cell Carcinoma.Front Genet. 2022 Apr 11;13:767166. doi: 10.3389/fgene.2022.767166. eCollection 2022. Front Genet. 2022. PMID: 35480305 Free PMC article.
-
Identification of Novel Biomarkers for Predicting Prognosis and Immunotherapy Response in Head and Neck Squamous Cell Carcinoma Based on ceRNA Network and Immune Infiltration Analysis.Biomed Res Int. 2021 Dec 6;2021:4532438. doi: 10.1155/2021/4532438. eCollection 2021. Biomed Res Int. 2021. PMID: 34917682 Free PMC article.
-
Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks.Bioengineered. 2021 Dec;12(2):12821-12838. doi: 10.1080/21655979.2021.2003925. Bioengineered. 2021. PMID: 34898376 Free PMC article.
Cited by
-
Progress in Precision Medicine for Head and Neck Cancer.Cancers (Basel). 2024 Nov 4;16(21):3716. doi: 10.3390/cancers16213716. Cancers (Basel). 2024. PMID: 39518152 Free PMC article. Review.
-
A novel fumaric acid metabolism-related prognostic signature associated with prognosis and immune infiltration landscape in laryngeal squamous cell carcinoma.Transl Cancer Res. 2025 Jul 30;14(7):3991-4008. doi: 10.21037/tcr-2025-29. Epub 2025 Jul 25. Transl Cancer Res. 2025. PMID: 40792153 Free PMC article.
References
-
- Travassos D. C., Fernandes D., Massucato E. M. S., Navarro C. M., Bufalino A. Squamous cell carcinoma antigen as a prognostic marker and its correlation with clinicopathological features in head and neck squamous cell carcinoma: systematic review and meta-analysis. Journal of Oral Pathology & Medicine . 2018;47(1):3–10. doi: 10.1111/jop.12600. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

