Hyperspectral Technique Combined With Deep Learning Algorithm for Prediction of Phenotyping Traits in Lettuce
- PMID: 35845657
- PMCID: PMC9279906
- DOI: 10.3389/fpls.2022.927832
Hyperspectral Technique Combined With Deep Learning Algorithm for Prediction of Phenotyping Traits in Lettuce
Abstract
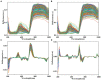
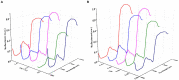
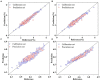
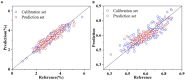
The currently available methods for evaluating most biochemical traits of plant phenotyping are destructive and have extremely low throughput. However, hyperspectral techniques can non-destructively obtain the spectral reflectance characteristics of plants, which can provide abundant biophysical and biochemical information. Therefore, plant spectra combined with machine learning algorithms can be used to predict plant phenotyping traits. However, the raw spectral reflectance characteristics contain noise and redundant information, thus can easily affect the robustness of the models developed via multivariate analysis methods. In this study, two end-to-end deep learning models were developed based on 2D convolutional neural networks (2DCNN) and fully connected neural networks (FCNN; Deep2D and DeepFC, respectively) to rapidly and non-destructively predict the phenotyping traits of lettuces from spectral reflectance. Three linear and two nonlinear multivariate analysis methods were used to develop models to weigh the performance of the deep learning models. The models based on multivariate analysis methods require a series of manual feature extractions, such as pretreatment and wavelength selection, while the proposed models can automatically extract the features in relation to phenotyping traits. A visible near-infrared hyperspectral camera was used to image lettuce plants growing in the field, and the spectra extracted from the images were used to train the network. The proposed models achieved good performance with a determination coefficient of prediction ( ) of 0.9030 and 0.8490 using Deep2D for soluble solids content and DeepFC for pH, respectively. The performance of the deep learning models was compared with five multivariate analysis method. The quantitative analysis showed that the deep learning models had higher than all the multivariate analysis methods, indicating better performance. Also, wavelength selection and different pretreatment methods had different effects on different multivariate analysis methods, and the selection of appropriate multivariate analysis methods and pretreatment methods increased more time and computational cost. Unlike multivariate analysis methods, the proposed deep learning models did not require any pretreatment or dimensionality reduction and thus are more suitable for application in high-throughput plant phenotyping platforms. These results indicate that the deep learning models can better predict phenotyping traits of plants using spectral reflectance.
Keywords: SSC; deep learning; hyperspectral imaging; lettuce; pH; plant phenotyping.
Copyright © 2022 Yu, Fan, Lu, Wen, Shao, Guo and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Aasen H., Burkart A., Bolten A., Bareth G. (2015). Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS J. Photogramm. Remote Sens. 108, 245–259. doi: 10.1016/j.isprsjprs.2015.08.002 - DOI
-
- Aich S., Stavness I.. (2017). Leaf counting with deep convolutional and deconvolutional networks. Proceedings of the IEEE International Conference on Computer Vision Workshops. IEEE. 2080–2089. doi: 10.7780/kjrs.2021.37.3.12 - DOI
-
- Chung J., Son M., Lee Y., Kim S. (2021). Estimation of soil moisture using Sentinel-1 SAR images and multiple linear regression model considering antecedent precipitations. Korean J. Remote Sens. 37, 515–530.
-
- De Corato U. (2020). Improving the shelf-life and quality of fresh and minimally-processed fruits and vegetables for a modern food industry: a comprehensive critical review from the traditional technologies into the most promising advancements. Crit. Rev. Food Sci. Nutr. 60, 940–975. doi: 10.1080/10408398.2018.1553025, PMID: - DOI - PubMed
-
- ElMasry G., Wang N., ElSayed A., Ngadi M. (2007). Hyperspectral imaging for nondestructive determination of some quality attributes for strawberry. J. Food Eng. 81, 98–107. doi: 10.1016/j.jfoodeng.2006.10.016 - DOI
LinkOut - more resources
Full Text Sources

