Reno-Protective Effect of Low Protein Diet Supplemented With α-Ketoacid Through Gut Microbiota and Fecal Metabolism in 5/6 Nephrectomized Mice
- PMID: 35845811
- PMCID: PMC9280408
- DOI: 10.3389/fnut.2022.889131
Reno-Protective Effect of Low Protein Diet Supplemented With α-Ketoacid Through Gut Microbiota and Fecal Metabolism in 5/6 Nephrectomized Mice
Abstract
Background: Low protein supplemented with α-ketoacid diet (LKD) was recommended to be an essential intervention to delay the progression of chronic kidney disease (CKD) in patients who were not yet on dialysis. Aberrant gut microbiota and metabolism have been reported to be highly associated with CKD. However, the effect of LKD on gut microbiota and related fecal metabolism in CKD remains unclear.
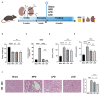
Methods: Mice were fed with normal protein diet (NPD group), low protein diet (LPD group), and low protein diet supplemented with α-ketoacid (LKD group) after 5/6 nephrectomy. At the end of the study, blood, kidney tissues, and feces were collected for biochemical analyses, histological, 16S rRNA sequence of gut microbiome, and untargeted fecal metabolomic analyses.
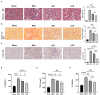
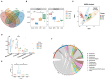
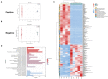
Results: Both LKD and LPD alleviate renal failure and fibrosis, and inflammatory statement in 5/6 nephrectomized mice, especially the LKD. In terms of gut microbiome, LKD significantly improved the dysbiosis induced by 5/6Nx, representing increased α-diversity and decreased F/B ratio. Compared with NPD, LKD significantly increased the abundance of g_Parasutterella, s_Parabacteroides_sp_CT06, f_Erysipelotrichaceae, g_Akkermansia, g_Gordonibacter, g_Faecalitalea, and s_Mucispirillum_sp_69, and decreased s_Lachnospiraceae_bacterium_28-4 and g_Lachnoclostridium. Moreover, 5/6Nx and LKD significantly altered fecal metabolome. Then, multi-omics analysis revealed that specific metabolites involved in glycerophospholipid, purine, vitamin B6, sphingolipid, phenylalanine, tyrosine and tryptophan biosynthesis, and microbes associated with LKD were correlated with the amelioration of CKD.
Conclusion: LKD had a better effect than LPD on delaying renal failure in 5/6 nephrectomy-induced CKD, which may be due to the regulation of affecting the gut microbiome and fecal metabolic profiles.
Keywords: 5/6Nx mice; chronic kidney disease; fecal metabolism; gut microbiota; low-protein diet supplemented with α-ketoacid; renal fibrosis.
Copyright © 2022 Zhu, He, Tang, Peng, Hu, Sun, Liu, Jin and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Microbiome-Metabolomics Analysis Reveals the Protection Mechanism of α-Ketoacid on Adenine-Induced Chronic Kidney Disease in Rats.Front Pharmacol. 2021 May 11;12:657827. doi: 10.3389/fphar.2021.657827. eCollection 2021. Front Pharmacol. 2021. PMID: 34045965 Free PMC article.
-
Fecal metabonomics combined with 16S rRNA gene sequencing to analyze the changes of gut microbiota in rats with kidney-yang deficiency syndrome and the intervention effect of You-gui pill.J Ethnopharmacol. 2019 Nov 15;244:112139. doi: 10.1016/j.jep.2019.112139. Epub 2019 Aug 8. J Ethnopharmacol. 2019. PMID: 31401318
-
Disrupted gut microbiota promotes the progression of chronic kidney disease in 5/6 nephrectomy mice by Bacillus pumilus gavage.Front Cell Infect Microbiol. 2025 Mar 18;15:1548767. doi: 10.3389/fcimb.2025.1548767. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40171160 Free PMC article.
-
Longitudinal analysis of fecal microbiome and metabolome during renal fibrotic progression in a unilateral ureteral obstruction animal model.Eur J Pharmacol. 2020 Nov 5;886:173555. doi: 10.1016/j.ejphar.2020.173555. Epub 2020 Sep 14. Eur J Pharmacol. 2020. PMID: 32937112
-
Effects of Low Protein Diet on Modulating Gut Microbiota in Patients with Chronic Kidney Disease: A Systematic Review and Meta-analysis of International Studies.Int J Med Sci. 2021 Oct 25;18(16):3839-3850. doi: 10.7150/ijms.66451. eCollection 2021. Int J Med Sci. 2021. PMID: 34790060 Free PMC article.
Cited by
-
Comparative Analysis of Gut Microbiomes in Laboratory Chinchillas, Ferrets, and Marmots: Implications for Pathogen Infection Research.Microorganisms. 2024 Mar 24;12(4):646. doi: 10.3390/microorganisms12040646. Microorganisms. 2024. PMID: 38674591 Free PMC article.
-
Genetic evidence supporting the causal role of gut microbiota in chronic kidney disease and chronic systemic inflammation in CKD: a bilateral two-sample Mendelian randomization study.Front Immunol. 2023 Nov 2;14:1287698. doi: 10.3389/fimmu.2023.1287698. eCollection 2023. Front Immunol. 2023. PMID: 38022507 Free PMC article.
-
Metabolomic analysis of rumen fluid in Tan sheep reveals sex-specific key metabolites and pathways associated with residual feed intake.Sci Rep. 2025 Jul 1;15(1):22115. doi: 10.1038/s41598-025-06182-8. Sci Rep. 2025. PMID: 40594387 Free PMC article.
-
Causal relationship between gut microbiota and kidney diseases: a two-sample Mendelian randomization study.Front Immunol. 2024 Jan 12;14:1277554. doi: 10.3389/fimmu.2023.1277554. eCollection 2023. Front Immunol. 2024. PMID: 38283353 Free PMC article.
References
LinkOut - more resources
Full Text Sources

