Polymorphisms and AR: A Systematic Review and Meta-Analyses
- PMID: 35846137
- PMCID: PMC9284009
- DOI: 10.3389/fgene.2022.899923
Polymorphisms and AR: A Systematic Review and Meta-Analyses
Abstract
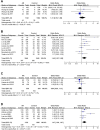
Background: Allergic rhinitis (AR) is an especially common disorder associated with both environmental and genetic factors, and a lot of researchers have attempted to find polymorphisms which predisposed to the disease. We conducted a meta-analysis of the most frequently researched polymorphisms to find those genes which may be susceptible to AR and then may be of value in diagnosis. Methods: Pubmed and China National Knowledge Infrastructure (CNKI) databases were searched to screen out eligible studies focusing on the correlation between polymorphisms and AR susceptibility, and then polymorphisms cited in at least 3 studies were selected. Results: The 142 papers originally selected cited 78 genes. Twelve genes (coinciding with 23 polymorphisms) were reported in more than three papers. Twenty-three polymorphisms were involved in the meta-analysis. Among the 23 polymorphisms, only 4 were found to be related to the risk of AR: IL-13 rs20541, CTLA-4 rs11571302, IL-4R RS1801275 and ACE (I/D). The remaining 19 of the 23 polymorphisms were not associated with AR. Conclusion: We found polymorphisms that could be used for AR diagnosing and those that were unrelated to AR. This may be the first step in detecting polymorphic combinations susceptible to AR (IL-13 RS20541, CTLA-4 RS11571302, IL-4R RS1801275 and ACE (I/D). In addition, our results may improve AR diagnosis and contribute to the intensive study of AR.
Keywords: ACE; CTLA-4; IL-13; IL-4R; allergic rhinitis; meta-analysis; polymorphism.
Copyright © 2022 Xiang, Zeng, Wang, Yang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
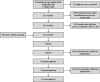

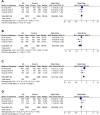
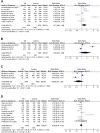
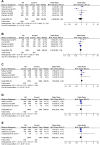

Similar articles
-
Polymorphisms and NIHL: a systematic review and meta-analyses.Front Cell Neurosci. 2023 Jun 15;17:1175427. doi: 10.3389/fncel.2023.1175427. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37396925 Free PMC article.
-
The Role of Interleukin-4 and 13 Gene Polymorphisms in Allergic Rhinitis: A Case Control Study.Rep Biochem Mol Biol. 2019 Jul;8(2):111-118. Rep Biochem Mol Biol. 2019. PMID: 31832433 Free PMC article.
-
[Association of CTLA-4 gene polymorphism in allergic rhinitis with asthma or not in children].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2016 Oct 20;30(20):1597-1600. doi: 10.13201/j.issn.1001-1781.2016.20.004. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2016. PMID: 29871152 Chinese.
-
Gene polymorphisms of Interleukin-4 in allergic rhinitis and its association with clinical phenotypes.Am J Otolaryngol. 2013 Nov-Dec;34(6):676-81. doi: 10.1016/j.amjoto.2013.05.002. Epub 2013 Sep 26. Am J Otolaryngol. 2013. PMID: 24075353
-
Prevalence of allergic rhinitis comorbidity with asthma and asthma with allergic rhinitis in China: A meta-analysis.Asian Pac J Allergy Immunol. 2019 Dec;37(4):220-225. doi: 10.12932/AP-120417-0072. Asian Pac J Allergy Immunol. 2019. PMID: 30525742
Cited by
-
Genetic association of beta-lactams-induced hypersensitivity reactions: A systematic review of genome-wide evidence and meta-analysis of candidate genes.World Allergy Organ J. 2023 Sep 22;16(9):100816. doi: 10.1016/j.waojou.2023.100816. eCollection 2023 Sep. World Allergy Organ J. 2023. PMID: 37780578 Free PMC article.
-
Correlation between forkhead box P3 (rs3761548) gene polymorphism and serum interleukin13 as biomarkers of severity in Egyptian allergic conjunctivitis: a retrospective study.Front Allergy. 2024 Sep 25;5:1437600. doi: 10.3389/falgy.2024.1437600. eCollection 2024. Front Allergy. 2024. PMID: 39386093 Free PMC article.
-
Association of Genetic Variations in The PIK3-AKT-mTOR Pathway with Endometriosis Susceptibility: A Preliminary Case-Control Study.Int J Fertil Steril. 2025 Mar 11;19(2):164-171. doi: 10.22074/ijfs.2024.2015384.1567. Int J Fertil Steril. 2025. PMID: 40200774 Free PMC article.
References
-
- Baldini M., Carla Lohman I., Halonen M., Erickson R. P., Holt P. G., Martinez F. D. (1999). A Polymorphism* in the 5 ′ Flanking Region of the CD14 Gene Is Associated with Circulating Soluble CD14 Levels and with Total Serum Immunoglobulin E. Am. J. Respir. Cell Mol. Biol. 20 (5), 976–983. 10.1165/ajrcmb.20.5.3494 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

