Multiple Novel Mosquito-Borne Zoonotic Viruses Revealed in Pangolin Virome
- PMID: 35846764
- PMCID: PMC9277073
- DOI: 10.3389/fcimb.2022.874003
Multiple Novel Mosquito-Borne Zoonotic Viruses Revealed in Pangolin Virome
Abstract
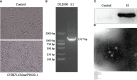
Swab samples were collected from 34 pangolins in Guangxi Province, China. Metavirome sequencing and bioinformatics approaches were undertaken to determine the abundant viral sequences in the viromes. The results showed that the viral sequences belong to 24 virus taxonomic families. To verify the results, PCR combined with phylogenetic analysis was conducted. Some viral sequences including Japanese encephalitis virus (JEV), Getah virus (GETV), and chikungunya virus (CHIKV) were detected. On the basis of the metavirome analysis, seven segments belonging to JEV were further identified through PCR amplification. Sequence comparison showed that, among seven sequences, JEV-China/P2020E-1 displayed the highest nucleotide (80.6%), with the JEV isolated in South Korea, 1988, and all of which belonging to genotype III. Seven CHIKV sequences were detected, with the highest homology (80.6%) to the Aedes africanus in Côte d'Ivoire, 1993. Moreover, passage from BHK-21 to Vero cells makes the newly isolated CHIKV-China/P2020-1 more contagious. In addition, the newly verified GETV sequences shared 86.4% identity with the 1955 GETV isolated from Malaysia. Some sudden and recurrent viruses have also been observed from the virome of pangolin in Guangxi Province, China; hence, dissemination tests will be implemented in the future.
Keywords: metagenomic analysis; mosquito-borne virus; pangolin; phylogenetic analysis; viral isolation and identification.
Copyright © 2022 Zhang, Zheng, Zhang, Feng, Peng, Li, Li, Zhang, Li and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Cabral-Castro M. J., Peralta R., Cavalcanti M. G., Puccioni-Sohler M., Carvalho V. L., da Costa Vasconcelos P. F., et al. . (2016). A Luminex-Based Single DNA Fragment Amplification Assay as a Practical Tool for Detecting and Serotyping Dengue Virus. J. Virol. Methods 236, 18–24. doi: 10.1016/j.jviromet.2016.07.003 - DOI - PubMed
-
- Guanrong F., Jinyong Z., Ying Z., Chenghui L., Duo Z., Yiquan L., et al. . (2022). Metagenomic Analysis of Togaviridae in Mosquito Viromes Isolated From Yunnan Province in China Reveals Genes From Chikungunya and Ross River Viruses. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.849662 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

