Development of Bovine Gastric Organoids as a Novel In Vitro Model to Study Host-Parasite Interactions in Gastrointestinal Nematode Infections
- PMID: 35846775
- PMCID: PMC9281477
- DOI: 10.3389/fcimb.2022.904606
Development of Bovine Gastric Organoids as a Novel In Vitro Model to Study Host-Parasite Interactions in Gastrointestinal Nematode Infections
Abstract
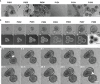
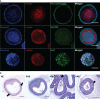
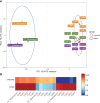
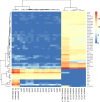
Gastro-intestinal nematode (GIN) parasites are a major cause of production losses in grazing cattle, primarily through reduced growth rates in young animals. Control of these parasites relies heavily on anthelmintic drugs; however, with growing reports of resistance to currently available anthelmintics, alternative methods of control are required. A major hurdle in this work has been the lack of physiologically relevant in vitro infection models that has made studying precise interactions between the host and the GINs difficult. Such mechanistic insights into the infection process will be valuable for the development of novel targets for drugs, vaccines, or other interventions. Here we created bovine gastric epithelial organoids from abomasal gastric tissue and studied their application as in vitro models for understanding host invasion by GIN parasites. Transcriptomic analysis of gastric organoids across multiple passages and the corresponding abomasal tissue showed conserved expression of tissue-specific genes across samples, demonstrating that the organoids are representative of bovine gastric tissue from which they were derived. We also show that self-renewing and self-organising three-dimensional organoids can also be serially passaged, cryopreserved, and resuscitated. Using Ostertagia ostertagi, the most pathogenic gastric parasite in cattle in temperate regions, we show that cattle gastric organoids are biologically relevant models for studying GIN invasion in the bovine abomasum. Within 24 h of exposure, exsheathed larvae rapidly and repeatedly infiltrated the lumen of the organoids. Prior to invasion by the parasites, the abomasal organoids rapidly expanded, developing a 'ballooning' phenotype. Ballooning of the organoids could also be induced in response to exposure to parasite excretory/secretory products. In summary, we demonstrate the power of using abomasal organoids as a physiologically relevant in vitro model system to study interactions of O. ostertagi and other GIN with bovine gastrointestinal epithelium.
Keywords: Ostertagia ostertagi; gastrointestinal; host-pathogen interactions; nematodes; three-dimensional (3D) organoids; tissue remodelling.
Copyright © 2022 Faber, Smith, Price, Steele, Hildersley, Morrison, Mabbott, Nisbet and McNeilly.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This study received funding from Zoetis. The funder had the following involvement with the study: the decision to submit the manuscript for publication.
Figures


Similar articles
-
Novel epidemiological model of gastrointestinal nematode infection to assess grazing cattle resilience by integrating host growth, parasite, grass and environmental dynamics.Int J Parasitol. 2023 Mar;53(3):133-155. doi: 10.1016/j.ijpara.2022.11.009. Epub 2023 Jan 24. Int J Parasitol. 2023. PMID: 36706804
-
Role of the bovine immune system and genome in resistance to gastrointestinal nematodes.Vet Parasitol. 2001 Jul 12;98(1-3):51-64. doi: 10.1016/s0304-4017(01)00423-x. Vet Parasitol. 2001. PMID: 11516579 Review.
-
The identification of small molecule inhibitors with anthelmintic activities that target conserved proteins among ruminant gastrointestinal nematodes.mBio. 2024 Mar 13;15(3):e0009524. doi: 10.1128/mbio.00095-24. Epub 2024 Feb 15. mBio. 2024. PMID: 38358246 Free PMC article.
-
Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi.Int J Mol Sci. 2025 Mar 4;26(5):2264. doi: 10.3390/ijms26052264. Int J Mol Sci. 2025. PMID: 40076885 Free PMC article.
-
Effects of gastrointestinal nematode infection on the ruminant immune system.Vet Parasitol. 1997 Nov;72(3-4):327-37; discussion 337-43. doi: 10.1016/s0304-4017(97)00104-0. Vet Parasitol. 1997. PMID: 9460205 Review.
Cited by
-
Characterization of bovine and ovine basal-out and apical-out ileum organoids.R Soc Open Sci. 2025 Jul 30;12(7):250326. doi: 10.1098/rsos.250326. eCollection 2025 Jul. R Soc Open Sci. 2025. PMID: 40740711 Free PMC article.
-
A secreted helminth microRNA suppresses gastrointestinal cell differentiation required for innate immunity.Front Immunol. 2025 Mar 27;16:1558132. doi: 10.3389/fimmu.2025.1558132. eCollection 2025. Front Immunol. 2025. PMID: 40213548 Free PMC article.
-
Development of Sheep Intestinal Organoids for Studying Deoxynivalenol-Induced Toxicity.Int J Mol Sci. 2025 Jan 23;26(3):955. doi: 10.3390/ijms26030955. Int J Mol Sci. 2025. PMID: 39940725 Free PMC article.
-
In vitro co-culture of Fasciola hepatica newly excysted juveniles (NEJs) with 3D HepG2 spheroids permits novel investigation of host-parasite interactions.Virulence. 2025 Dec;16(1):2482159. doi: 10.1080/21505594.2025.2482159. Epub 2025 Mar 25. Virulence. 2025. PMID: 40132201 Free PMC article.
-
Equine enteroid-derived monolayers recapitulate key features of parasitic intestinal nematode infection.Vet Res. 2024 Feb 27;55(1):25. doi: 10.1186/s13567-024-01266-1. Vet Res. 2024. PMID: 38414039 Free PMC article.
References
-
- Albuquerque A. C. A., Bassetto C. C., Almeida F. A., Hildersley K. A., McNeilly T. N., Britton C., et al. . (2019). Differences in Immune Responses to Haemonchus Contortus Infection in the Susceptible Ile De France and the Resistant Santa Ines Sheep Under Different Anthelmintic Treatments Regimens. Vet. Res. 50, 104. doi: 10.1186/s13567-019-0722-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

