Dynamics of Mismatch and Alternative Excision-Dependent Repair in Replicating Bacillus subtilis DNA Examined Under Conditions of Neutral Selection
- PMID: 35847079
- PMCID: PMC9280176
- DOI: 10.3389/fmicb.2022.866089
Dynamics of Mismatch and Alternative Excision-Dependent Repair in Replicating Bacillus subtilis DNA Examined Under Conditions of Neutral Selection
Abstract
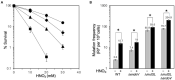
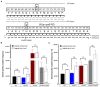
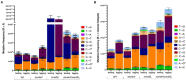
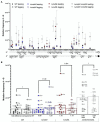
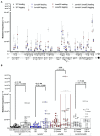
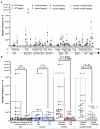
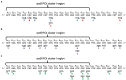
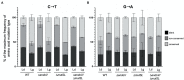
Spontaneous DNA deamination is a potential source of transition mutations. In Bacillus subtilis, EndoV, a component of the alternative excision repair pathway (AER), counteracts the mutagenicity of base deamination-induced mispairs. Here, we report that the mismatch repair (MMR) system, MutSL, prevents the harmful effects of HNO2, a deaminating agent of Cytosine (C), Adenine (A), and Guanine (G). Using Maximum Depth Sequencing (MDS), which measures mutagenesis under conditions of neutral selection, in B. subtilis strains proficient or deficient in MutSL and/or EndoV, revealed asymmetric and heterogeneous patterns of mutations in both DNA template strands. While the lagging template strand showed a higher frequency of C → T substitutions; G → A mutations, occurred more frequently in the leading template strand in different genetic backgrounds. In summary, our results unveiled a role for MutSL in preventing the deleterious effects of base deamination and uncovered differential patterns of base deamination processing by the AER and MMR systems that are influenced by the sequence context and the replicating DNA strand.
Keywords: AER repair; DNA mispairs; base deamination; maximum depth sequencing; mismatch repair.
Copyright © 2022 Patlán-Vázquez, Ayala-García, Vallin, Cortés, Vásquez-Morales, Robleto, Nudler and Pedraza-Reyes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ayala-García V. M., Valenzuela-García L. I., Setlow P., Pedraza-Reyes M. (2016). Aag hypoxanthine-DNA glycosylase is synthesized in the forespore compartment and involved in counteracting the genotoxic and mutagenic effects of hypoxanthine and alkylated bases in DNA during Bacillus subtilis sporulation. J. Bacteriol. 198, 3345–3354. doi: 10.1128/jb.00625-16, PMID: - DOI - PMC - PubMed
-
- Castañeda-García A., Martín-Blecua I., Cebrián-Sastre E., Chiner-Oms A., Torres-Puente M., Comas I., et al. . (2020). Specificity and mutagenesis bias of the mycobacterial alternative mismatch repair analyzed by mutation accumulation studies. Sci. Adv. 6:eaay4453. doi: 10.1126/sciadv.aay4453, PMID: - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

