Metagenomic Investigation of Ticks From Kenyan Wildlife Reveals Diverse Microbial Pathogens and New Country Pathogen Records
- PMID: 35847110
- PMCID: PMC9283121
- DOI: 10.3389/fmicb.2022.932224
Metagenomic Investigation of Ticks From Kenyan Wildlife Reveals Diverse Microbial Pathogens and New Country Pathogen Records
Abstract
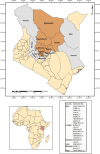
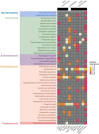
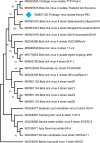
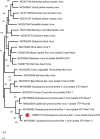
Focusing on the utility of ticks as xenosurveillance sentinels to expose circulating pathogens in Kenyan drylands, host-feeding ticks collected from wild ungulates [buffaloes, elephants, giraffes, hartebeest, impala, rhinoceros (black and white), zebras (Grévy's and plains)], carnivores (leopards, lions, spotted hyenas, wild dogs), as well as regular domestic and Boran cattle were screened for pathogens using metagenomics. A total of 75 host-feeding ticks [Rhipicephalus (97.3%) and Amblyomma (2.7%)] collected from 15 vertebrate taxa were sequenced in 46 pools. Fifty-six pathogenic bacterial species were detected in 35 pools analyzed for pathogens and relative abundances of major phyla. The most frequently observed species was Escherichia coli (62.8%), followed by Proteus mirabilis (48.5%) and Coxiella burnetii (45.7%). Francisella tularemia and Jingmen tick virus (JMTV) were detected in 14.2 and 13% of the pools, respectively, in ticks collected from wild animals and cattle. This is one of the first reports of JMTV in Kenya, and phylogenetic reconstruction revealed significant divergence from previously known isolates and related viruses. Eight fungal species with human pathogenicity were detected in 5 pools (10.8%). The vector-borne filarial pathogens (Brugia malayi, Dirofilaria immitis, Loa loa), protozoa (Plasmodium spp., Trypanosoma cruzi), and environmental and water-/food-borne pathogens (Entamoeba histolytica, Encephalitozoon intestinalis, Naegleria fowleri, Schistosoma spp., Toxoplasma gondii, and Trichinella spiralis) were detected. Documented viruses included human mastadenovirus C, Epstein-Barr virus and bovine herpesvirus 5, Trinbago virus, and Guarapuava tymovirus-like virus 1. Our findings confirmed that host-feeding ticks are an efficient sentinel for xenosurveillance and demonstrate clear potential for wildlife-livestock-human pathogen transfer in the Kenyan landscape.
Keywords: metagenomics; pathogen; surveillance; tick; wildlife.
Copyright © 2022 Ergunay, Mutinda, Bourke, Justi, Caicedo-Quiroga, Kamau, Mutura, Akunda, Cook, Gakuya, Omondi, Murray, Zimmerman and Linton.
Conflict of interest statement
MM was employed by Kenya Wildlife Services Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Molecular Detection of Tacheng Tick Virus-1 (TcTV-1) and Jingmen Tick Virus in Ticks Collected from Wildlife and Livestock in Turkey: First Indication of TcTV-1 Beyond China.Vector Borne Zoonotic Dis. 2023 Aug;23(8):419-427. doi: 10.1089/vbz.2023.0029. Epub 2023 Jun 9. Vector Borne Zoonotic Dis. 2023. PMID: 37294551
-
Ticks and prevalence of tick-borne pathogens from domestic animals in Ghana.Parasit Vectors. 2022 Mar 12;15(1):86. doi: 10.1186/s13071-022-05208-8. Parasit Vectors. 2022. PMID: 35279200 Free PMC article.
-
Pathogens, endosymbionts, and blood-meal sources of host-seeking ticks in the fast-changing Maasai Mara wildlife ecosystem.PLoS One. 2020 Aug 31;15(8):e0228366. doi: 10.1371/journal.pone.0228366. eCollection 2020. PLoS One. 2020. PMID: 32866142 Free PMC article.
-
Distribution and prevalence of ticks and tick-borne pathogens of wild animals in South Africa: A systematic review.Curr Res Parasitol Vector Borne Dis. 2022 Apr 26;2:100088. doi: 10.1016/j.crpvbd.2022.100088. eCollection 2022. Curr Res Parasitol Vector Borne Dis. 2022. PMID: 35601607 Free PMC article. Review.
-
Ticks and tick-borne diseases of bovines in a smallholder livestock context: The Pakistani example.Adv Parasitol. 2021;114:167-244. doi: 10.1016/bs.apar.2021.08.009. Epub 2021 Oct 9. Adv Parasitol. 2021. PMID: 34696843
Cited by
-
Jingmen tick virus: an emerging arbovirus with a global threat.mSphere. 2023 Oct 24;8(5):e0028123. doi: 10.1128/msphere.00281-23. Epub 2023 Sep 13. mSphere. 2023. PMID: 37702505 Free PMC article. Review.
-
Deciphering the microbial communities in ticks of Inner Mongolia: ecological determinants and pathogen profiles.Parasit Vectors. 2024 Nov 4;17(1):448. doi: 10.1186/s13071-024-06512-1. Parasit Vectors. 2024. PMID: 39497080 Free PMC article.
-
Hard ticks (Ixodida: Ixodidae) in the Colombian Caribbean harbor the Jingmen tick virus: an emerging arbovirus of public health concern.Parasit Vectors. 2024 Jun 25;17(1):268. doi: 10.1186/s13071-024-06362-x. Parasit Vectors. 2024. PMID: 38918818 Free PMC article.
-
Challenges and Opportunities in Pathogen Agnostic Sequencing for Public Health Surveillance: Lessons Learned From the Global Emerging Infections Surveillance Program.Health Secur. 2024 Jan-Feb;22(1):16-24. doi: 10.1089/hs.2023.0068. Epub 2023 Dec 6. Health Secur. 2024. PMID: 38054950 Free PMC article. No abstract available.
-
Canine Distemper Virus in Autochtonous and Imported Dogs, Southern Italy (2014-2021).Animals (Basel). 2022 Oct 20;12(20):2852. doi: 10.3390/ani12202852. Animals (Basel). 2022. PMID: 36290237 Free PMC article.
References
-
- Altschul S. F., Gish W., Millerm W., Myers E. W., Lipman D. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

