Humans and Hoofed Livestock Are the Main Sources of Fecal Contamination of Rivers Used for Crop Irrigation: A Microbial Source Tracking Approach
- PMID: 35847115
- PMCID: PMC9279616
- DOI: 10.3389/fmicb.2022.768527
Humans and Hoofed Livestock Are the Main Sources of Fecal Contamination of Rivers Used for Crop Irrigation: A Microbial Source Tracking Approach
Abstract
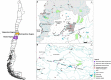
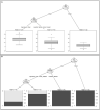
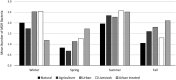
Freshwater bodies receive waste, feces, and fecal microorganisms from agricultural, urban, and natural activities. In this study, the probable sources of fecal contamination were determined. Also, antibiotic resistant bacteria (ARB) were detected in the two main rivers of central Chile. Surface water samples were collected from 12 sampling sites in the Maipo (n = 8) and Maule Rivers (n = 4) every 3 months, from August 2017 until April 2019. To determine the fecal contamination level, fecal coliforms were quantified using the most probable number (MPN) method and the source of fecal contamination was determined by Microbial Source Tracking (MST) using the Cryptosporidium and Giardia genotyping method. Separately, to determine if antimicrobial resistance bacteria (AMB) were present in the rivers, Escherichia coli and environmental bacteria were isolated, and the antibiotic susceptibility profile was determined. Fecal coliform levels in the Maule and Maipo Rivers ranged between 1 and 130 MPN/100-ml, and 2 and 30,000 MPN/100-ml, respectively. Based on the MST results using Cryptosporidium and Giardia host-specific species, human, cattle, birds, and/or dogs hosts were the probable sources of fecal contamination in both rivers, with human and cattle host-specific species being more frequently detected. Conditional tree analysis indicated that coliform levels were significantly associated with the river system (Maipo versus Maule), land use, and season. Fecal coliform levels were significantly (p < 0.006) higher at urban and agricultural sites than at sites immediately downstream of treatment centers, livestock areas, or natural areas. Three out of eight (37.5%) E. coli isolates presented a multidrug-resistance (MDR) phenotype. Similarly, 6.6% (117/1768) and 5.1% (44/863) of environmental isolates, in Maipo and Maule River showed and MDR phenotype. Efforts to reduce fecal discharge into these rivers should thus focus on agriculture and urban land uses as these areas were contributing the most and more frequently to fecal contamination into the rivers, while human and cattle fecal discharges were identified as the most likely source of this fecal contamination by the MST approach. This information can be used to design better mitigation strategies, thereby reducing the burden of waterborne diseases and AMR in Central Chile.
Keywords: Cryptosporidium; Giardia; antimicrobial resistance; fecal coliforms; microbial source tracking; water quality; waterborne pathogens.
Copyright © 2022 Díaz-Gavidia, Barría, Weller, Salgado-Caxito, Estrada, Araya, Vera, Smith, Kim, Moreno-Switt, Olivares-Pacheco and Adell.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Adell A. D., Smith W. A., Shapiro K., Melli A., Conrad P. A. (2014). Molecular epidemiology of Cryptosporidium spp. and Giardia spp. in mussels (Mytilus californianus) and California Sea lions (Zalophus californianus) from Central California. Appl. Environ. Microbiol. 80, 7732–7740. doi: 10.1128/AEM.02922-14, PMID: - DOI - PMC - PubMed
-
- Bambic D., McBride G., Miller W., Stott R., Wuertz S. (2011). Quantification of Pathogens and Sources of Microbial Indicators for QMRA in Recreational Waters. Vol. 10. IWA Publishing.
LinkOut - more resources
Full Text Sources
Miscellaneous

