Enterovirus A71 antivirals: Past, present, and future
- PMID: 35847514
- PMCID: PMC9279511
- DOI: 10.1016/j.apsb.2021.08.017
Enterovirus A71 antivirals: Past, present, and future
Abstract
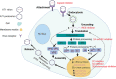
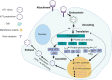
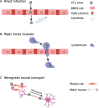
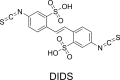
Enterovirus A71 (EV-A71) is a significant human pathogen, especially in children. EV-A71 infection is one of the leading causes of hand, foot, and mouth diseases (HFMD), and can lead to neurological complications such as acute flaccid myelitis (AFM) in severe cases. Although three EV-A71 vaccines are available in China, they are not broadly protective and have reduced efficacy against emerging strains. There is currently no approved antiviral for EV-A71. Significant progress has been made in developing antivirals against EV-A71 by targeting both viral proteins and host factors. However, viral capsid inhibitors and protease inhibitors failed in clinical trials of human rhinovirus infection due to limited efficacy or side effects. This review discusses major discoveries in EV-A71 antiviral development, analyzes the advantages and limitations of each drug target, and highlights the knowledge gaps that need to be addressed to advance the field forward.
Keywords: 2C protein; Acute flaccid myelitis; Antivirals; EV-A71; Enterovirus A71; Foot and mouth disease (HFMD); Hand; Picornavirus.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures
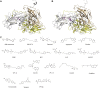
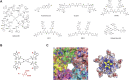
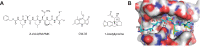

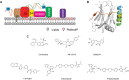

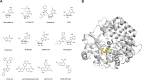


References
-
- Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

