Establishment of a reference single-cell RNA sequencing dataset for human pancreatic adenocarcinoma
- PMID: 35847558
- PMCID: PMC9283889
- DOI: 10.1016/j.isci.2022.104659
Establishment of a reference single-cell RNA sequencing dataset for human pancreatic adenocarcinoma
Abstract
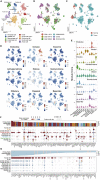
Single-cell RNA sequencing (scRNAseq) has been used to assess the intra-tumor heterogeneity and microenvironment of pancreatic ductal adenocarcinoma (PDAC). However, previous knowledge is not fully universalized. Here, we built a single cell atlas of PDAC from six datasets containing over 70 samples and >130,000 cells, and demonstrated its application to the reanalysis of the previous bulk transcriptomic cohorts and inferring cell-cell communications. The cell decomposition of bulk transcriptomics using scRNAseq data showed the cellular heterogeneity of PDAC; moreover, high levels of tumor cells and fibroblasts were indicative of poor-prognosis. Refined tumor subtypes signature indicated the tumor cell dynamics in intra-tumor and their specific regulatory network. We further identified functionally distinct tumor clusters that had close interaction with fibroblast subtypes via different signaling pathways dependent on subtypes. Our analysis provided a reference dataset for PDAC and showed its utility in research on the microenvironment of intra-tumor heterogeneity.
Keywords: Cancer; Cancer systems biology; Transcriptomics.
© 2022 The Author(s).
Conflict of interest statement
Partial institutional endowments were received from Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan), Hirotsu Bio Science Inc. (Tokyo, Japan), Kinshu-kai Medical Corporation (Osaka, Japan), Kyowa-kai Medical Corporation (Osaka, Japan), IDEA Consultants Inc. (Tokyo, Japan), and Unitech Co. Ltd. (Chiba, Japan).
Figures






References
-
- Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M., et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

