Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Diospyros oleifera, the first representative from the family Ebenaceae
- PMID: 35847622
- PMCID: PMC9283892
- DOI: 10.1016/j.heliyon.2022.e09870
Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Diospyros oleifera, the first representative from the family Ebenaceae
Abstract
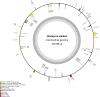
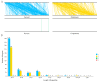
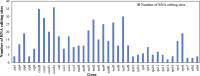
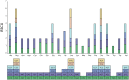
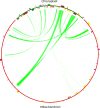
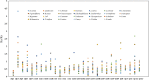
Plant mitochondrial genomes are a valuable source of genetic information for a better understanding of phylogenetic relationships. However, no mitochondrial genome of any species in Ebenaceae has been reported. In this study, we reported the first mitochondrial genome of an Ebenaceae model plant Diospyros oleifera. The mitogenome was 493,958 bp in length, contained 39 protein-coding genes, 27 transfer RNA genes, and 3 ribosomal RNA genes. The rps2 and rps11 genes were missing in the D. oleifera mt genome, while the rps10 gene was identified. The length of the repetitive sequence in the D. oleifera mt genome was 31 kb, accounting for 6.33%. A clear bias in RNA-editing sites were found in the D. oleifera mt genome. We also detected 28 chloroplast-derived fragments significantly associated with D. oleifera mt genes, indicating intracellular tRNA genes transferred frequently from chloroplasts to mitochondria in D. oleifera. Phylogenetic analysis based on the mt genomes of D. oleifera and 27 other taxa reflected the exact evolutionary and taxonomic status of D. oleifera. Ka/Ks analysis revealed that 95.16% of the protein-coding genes in the D. oleifera mt genome had undergone negative selections. But, the rearrangement of mitochondrial genes has been widely occur among D. oleifera and these observed species. These results will lay the foundation for identifying further evolutionary relationships within Ebenaceae.
Keywords: Diospyros oleifera; Mitochondrial genome; Phylogenetic analysis.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Photinia serratifolia.Sci Rep. 2023 Jan 14;13(1):770. doi: 10.1038/s41598-022-24327-x. Sci Rep. 2023. PMID: 36641495 Free PMC article.
-
Characterization and phylogenetic analysis of the chloroplast genome of Diospyros aff. oleifera.Mitochondrial DNA B Resour. 2024 Oct 28;9(11):1467-1472. doi: 10.1080/23802359.2024.2419451. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 39479478 Free PMC article.
-
A high-quality chromosomal genome assembly of Diospyros oleiferaCheng.Gigascience. 2020 Jan 1;9(1):giz164. doi: 10.1093/gigascience/giz164. Gigascience. 2020. PMID: 31944244 Free PMC article.
-
Five Complete Chloroplast Genome Sequences from Diospyros: Genome Organization and Comparative Analysis.PLoS One. 2016 Jul 21;11(7):e0159566. doi: 10.1371/journal.pone.0159566. eCollection 2016. PLoS One. 2016. PMID: 27442423 Free PMC article.
-
Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca.BMC Genomics. 2021 Mar 9;22(1):167. doi: 10.1186/s12864-021-07490-9. BMC Genomics. 2021. PMID: 33750312 Free PMC article.
Cited by
-
Structural and phylogenetic comparisons of the complete mitochondrial genomes among taxa in genus Toona.Sci Rep. 2025 Jun 25;15(1):20296. doi: 10.1038/s41598-025-06816-x. Sci Rep. 2025. PMID: 40562763 Free PMC article.
-
A High-Quality Assembly and Comparative Analysis of the Mitogenome of Actinidia macrosperma.Genes (Basel). 2024 Apr 19;15(4):514. doi: 10.3390/genes15040514. Genes (Basel). 2024. PMID: 38674448 Free PMC article.
-
Assembly and comparative analysis of the complete mitochondrial genome of Brassica rapa var. Purpuraria.BMC Genomics. 2024 Jun 1;25(1):546. doi: 10.1186/s12864-024-10457-1. BMC Genomics. 2024. PMID: 38824587 Free PMC article.
-
Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Lagenaria siceraria, a cucurbit crop.Front Plant Sci. 2025 Jul 22;16:1599596. doi: 10.3389/fpls.2025.1599596. eCollection 2025. Front Plant Sci. 2025. PMID: 40765853 Free PMC article.
-
Assembly and comparative analysis of the complete mitochondrial genome of Fritillaria ussuriensis Maxim. (Liliales: Liliaceae), an endangered medicinal plant.BMC Genomics. 2024 Aug 8;25(1):773. doi: 10.1186/s12864-024-10680-w. BMC Genomics. 2024. PMID: 39118028 Free PMC article.
References
-
- Fajardo D., Schlautman B., Steffan S., Polashock J., Vorsa N., Zalapa J. The American cranberry mitochondrial genome reveals the presence of selenocysteine (tRNA-Sec and SECIS) insertion machinery in land plants. Gene. 2014;536(2):336–343. - PubMed
-
- Vallejo-Marin M., Cooley A.M., Lee M.Y., Folmer M., McKain M.R., Puzey J.R. Strongly asymmetric hybridization barriers shape the origin of a new polyploid species and its hybrid ancestor. Am. J. Bot. 2016;103(7):1272–1288. - PubMed
-
- Greiner S., Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays. 2013;35(4):354–365. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

