Gut Microbiota in Systemic Lupus Erythematosus and Correlation With Diet and Clinical Manifestations
- PMID: 35847775
- PMCID: PMC9279557
- DOI: 10.3389/fmed.2022.915179
Gut Microbiota in Systemic Lupus Erythematosus and Correlation With Diet and Clinical Manifestations
Abstract
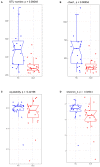
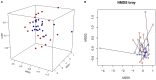
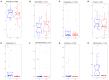
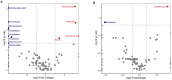
Despite the existing studies relating systemic lupus erythematosus (SLE) to changes in gut microbiota, the latter is affected by external factors such as diet and living environment. Herein, we compared the diversity and composition of gut microbiota in SLE patients and in their healthy family members who share the same household, to link gut microbiota, diet and SLE clinical manifestations. The study cohort included 19 patients with SLE and 19 of their healthy family members. Daily nutrition was assessed using a food frequency questionnaire (FFQ). Microbiota was analyzed using amplicons from the V4 regions of the 16S rRNA gene, to obtain microbiota diversity, taxa relative abundances and network analysis. The gut microbiota in the SLE group had lower alpha diversity and higher heterogeneity than the control group. SLE patients had decreased Acidobacteria, Gemmatimonadetes, Nitrospirae and Planctomycetes at the phylum level, and increased Streptococcus, Veillonella, Clostridium_XI, and Rothia at the genus level. Streptococcus was extremely enriched among patients with lupus nephritis. Lactobacillus, Clostridium_XlVa, Lachnospiracea_incertae_sedis and Parasutterella OTUs were associated with diet and clinical features of SLE. Finally, the gut microbiota of SLE patients remained different from that in healthy controls even after accounting for living conditions and diet.
Keywords: Diets; Gut microbiota; Network analysis; Streptococcus; Systemic lupus erythematosus.
Copyright © 2022 Wang, Shu, Song, Liu, Qu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Proton pump inhibitors induce changes in the gut microbiome composition of systemic lupus erythematosus patients.BMC Microbiol. 2022 Apr 27;22(1):117. doi: 10.1186/s12866-022-02533-x. BMC Microbiol. 2022. PMID: 35477382 Free PMC article.
-
Altered gut fungi in systemic lupus erythematosus - A pilot study.Front Microbiol. 2022 Dec 5;13:1031079. doi: 10.3389/fmicb.2022.1031079. eCollection 2022. Front Microbiol. 2022. PMID: 36545195 Free PMC article.
-
Systemic lupus erythematosus patients have a distinct structural and functional skin microbiota compared with controls.Lupus. 2021 Sep;30(10):1553-1564. doi: 10.1177/09612033211025095. Epub 2021 Jun 18. Lupus. 2021. PMID: 34139926
-
Altered Composition of Gut Microbiota in Depression: A Systematic Review.Front Psychiatry. 2020 Jun 10;11:541. doi: 10.3389/fpsyt.2020.00541. eCollection 2020. Front Psychiatry. 2020. PMID: 32587537 Free PMC article.
-
A Scoping Review of the Positive and Negative Bacteria Associated With the Gut Microbiomes of Systemic Lupus Erythematosus Patients.Cureus. 2024 Apr 3;16(4):e57512. doi: 10.7759/cureus.57512. eCollection 2024 Apr. Cureus. 2024. PMID: 38707123 Free PMC article.
Cited by
-
The Disease with a Thousand Faces and the Human Microbiome-A Physiopathogenic Intercorrelation in Pediatric Practice.Nutrients. 2023 Jul 28;15(15):3359. doi: 10.3390/nu15153359. Nutrients. 2023. PMID: 37571295 Free PMC article. Review.
-
Racial Differences in Plasma Microbial Translocation and Plasma Microbiome, Implications in Systemic Lupus Erythematosus Disease Pathogenesis.ACR Open Rheumatol. 2024 Jun;6(6):365-374. doi: 10.1002/acr2.11664. Epub 2024 Apr 2. ACR Open Rheumatol. 2024. PMID: 38563441 Free PMC article.
-
Alterations of the gut microbiota in the lupus nephritis: a systematic review.Ren Fail. 2023;45(2):2285877. doi: 10.1080/0886022X.2023.2285877. Epub 2023 Nov 23. Ren Fail. 2023. PMID: 37994423 Free PMC article.
-
Causal relationship between gut microbiome, immune cell, and systemic lupus erythematosus: A Mendelian randomization analysis.Medicine (Baltimore). 2025 Aug 1;104(31):e43703. doi: 10.1097/MD.0000000000043703. Medicine (Baltimore). 2025. PMID: 40760547 Free PMC article.
-
Differential impact of environmental factors on systemic and localized autoimmunity.Front Immunol. 2023 May 22;14:1147447. doi: 10.3389/fimmu.2023.1147447. eCollection 2023. Front Immunol. 2023. PMID: 37283765 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

