Mortality Prediction in Sepsis With an Immune-Related Transcriptomics Signature: A Multi-Cohort Analysis
- PMID: 35847809
- PMCID: PMC9280291
- DOI: 10.3389/fmed.2022.930043
Mortality Prediction in Sepsis With an Immune-Related Transcriptomics Signature: A Multi-Cohort Analysis
Abstract
Background: Novel biomarkers are needed to progress toward individualized patient care in sepsis. The immune profiling panel (IPP) prototype has been designed as a fully-automated multiplex tool measuring expression levels of 26 genes in sepsis patients to explore immune functions, determine sepsis endotypes and guide personalized clinical management. The performance of the IPP gene set to predict 30-day mortality has not been extensively characterized in heterogeneous cohorts of sepsis patients.
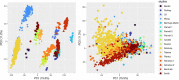
Methods: Publicly available microarray data of sepsis patients with widely variable demographics, clinical characteristics and ethnical background were co-normalized, and the performance of the IPP gene set to predict 30-day mortality was assessed using a combination of machine learning algorithms.
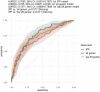
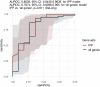
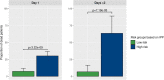
Results: We collected data from 1,801 arrays sampled on sepsis patients and 598 sampled on controls in 17 studies. When gene expression was assayed at day 1 following admission (1,437 arrays sampled on sepsis patients, of whom 1,161 were alive and 276 (19.2%) were dead at day 30), the IPP gene set showed good performance to predict 30-day mortality, with an area under the receiving operating characteristics curve (AUROC) of 0.710 (CI 0.652-0.768). Importantly, there was no statistically significant improvement in predictive performance when training the same models with all genes common to the 17 microarray studies (n = 7,122 genes), with an AUROC = 0.755 (CI 0.697-0.813, p = 0.286). In patients with gene expression data sampled at day 3 following admission or later, the IPP gene set had higher performance, with an AUROC = 0.804 (CI 0.643-0.964), while the total gene pool had an AUROC = 0.787 (CI 0.610-0.965, p = 0.811).
Conclusion: Using pooled publicly-available gene expression data from multiple cohorts, we showed that the IPP gene set, an immune-related transcriptomics signature conveys relevant information to predict 30-day mortality when sampled at day 1 following admission. Our data also suggests that higher predictive performance could be obtained when assaying gene expression at later time points during the course of sepsis. Prospective studies are needed to confirm these findings using the IPP gene set on its dedicated measurement platform.
Keywords: biomarker discovery; gene expression analysis; mortality; predictive modeling; sepsis; transcriptomics.
Copyright © 2022 Kreitmann, Bodinier, Fleurie, Imhoff, Cazalis, Peronnet, Cerrato, Tardiveau, Conti, Llitjos, Textoris, Monneret, Blein and Brengel-Pesce.
Conflict of interest statement
The IPP gene set has been filed for patent protection. LK was employed by, and has received research funding by bioMérieux. MB, AF, KI, M-AC, EP, EC, CT, J-FL, JT, SB, and KB-P were employed by bioMérieux. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

