Proteomic Analysis Identifies p62/SQSTM1 as a Critical Player in PARP Inhibitor Resistance
- PMID: 35847859
- PMCID: PMC9277186
- DOI: 10.3389/fonc.2022.908603
Proteomic Analysis Identifies p62/SQSTM1 as a Critical Player in PARP Inhibitor Resistance
Abstract
Poly (ADP-ribose) polymerase (PARP) inhibitors (PARPis) are currently being used for treating breast cancer patients with deleterious or suspected deleterious germline BRCA-mutated, HER2-negative locally advanced or metastatic diseases. Despite durable responses, almost all patients receiving PARPis ultimately develop resistance and succumb to their illness, but the mechanism of PARPi resistance is not fully understood. To better understand the mechanism of PARPi resistance, we established two olaparib-resistant SUM159 and MDA468 cells by chronically exposing olaparib-sensitive SUM159 and MDA468 cells to olaparib. Olaparib-resistant SUM159 and MDA468 cells displayed 5-fold and 7-fold more resistance over their corresponding counterparts. Despite defects in PARPi-induced DNA damage, these olaparib-resistant cells are sensitive to cisplatin-induced cell death. Using an unbiased proteomic approach, we identified 6 447 proteins, of which 107 proteins were differentially expressed between olaparib-sensitive and -resistant cells. Ingenuity pathway analysis (IPA) revealed a number of pathways that are significantly altered, including mTOR and ubiquitin pathways. Among these differentially expressed proteins, p62/SQSTM1 (thereafter p62), a scaffold protein, plays a critical role in binding to and delivering the ubiquitinated proteins to the autophagosome membrane for autophagic degradation, was significantly downregulated in olaparib-resistant cells. We found that autophagy inducers rapamycin and everolimus synergistically sensitize olaparib-resistant cells to olaparib. Moreover, p62 protein expression was correlated with better overall survival in estrogen receptor-negative breast cancer. Thus, these findings suggest that PARPi-sensitive TNBC cells hyperactivate autophagy as they develop acquired resistance and that pharmacological stimulation of excessive autophagy could lead to cell death and thus overcome PARPi resistance.
Keywords: PARP inhibitor; TNBC; autophagy; p62/SQSTM1; proteomics; rapamycin; resistance.
Copyright © 2022 Uddin, Zhou, Pimentel, Patrick, Kim, Shekhar and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


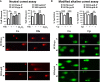
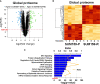


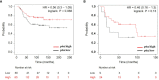
References
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

