miR-200a-3p- and miR-181-5p-Mediated HOXB5 Upregulation Promotes HCC Progression by Transcriptional Activation of EGFR
- PMID: 35847904
- PMCID: PMC9277860
- DOI: 10.3389/fonc.2022.822760
miR-200a-3p- and miR-181-5p-Mediated HOXB5 Upregulation Promotes HCC Progression by Transcriptional Activation of EGFR
Abstract
Background: Hepatocellular carcinoma (HCC) remains a worldwide burden. However, the mechanisms behind the malignant biological behavior of HCC remain unclear. The homeobox (HOX) family could act as either promoters or suppressors in different kinds of malignancies. Our study discovered the role of HOXB5 in regulating HCC progression.
Methods: The HOXB5 expression was assessed by RT-PCR analysis in human HCC samples and cell lines. HOXB5 transcriptional regulation of the EGFR was verified by the luciferase reporter assay and chromatin immunoprecipitation experiment. The oncogenic role of HOXB5 in HCC progression was analyzed by CCK8, colony-forming, and transwell assays.
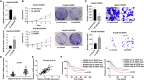
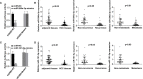
Results: Upregulation of HOXB5 was found in human HCC, and was strongly correlated with HCC tumor size, tumor-nodule metastasis, TNM stage, and relatively unfavorable OS and DFS. Ectopic expression of HOXB5 promoted the capacity of cell growth and clonogenicity, while the inhibition of HOXB5 decreased the proliferation and clonogenicity potential in vitro by CCK8 and colony-forming assays. In addition, HOXB5 also promoted cell migration by transwell experiment. Mechanism studies elucidated that HOXB5 triggers HCC progression via direct transcriptional activation of EGFR. The upregulation of HOXB5 is regulated by miR-200a-3p and miR-181-5p. Transfection of miR-200a-3p and miR-181-5p mimics blocked the cell proliferation and migration regulated by HOXB5, while overexpression of the 3'-UTR mutant HOXB5 abolished the suppressive effect of miR-200a-3p and miR-181-5p, but not the wild-type HOXB5.
Conclusion: HOXB5 is a promising prognostic factor in human HCC. Targeting miR-200a-3p and the miR-181-5p/HOXB5/EGFR signaling pathway may provide new options for the treatment strategies of HCC.
Keywords: EGFR; HCC; HOXB5; miR-181-5p; miR-200a-3p.
Copyright © 2022 Li, Li, Li, Ma, Liu, Kong and Xue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Homeobox B5 promotes metastasis and poor prognosis in Hepatocellular Carcinoma, via FGFR4 and CXCL1 upregulation.Theranostics. 2021 Mar 31;11(12):5759-5777. doi: 10.7150/thno.57659. eCollection 2021. Theranostics. 2021. PMID: 33897880 Free PMC article.
-
Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6.Cancer Cell Int. 2018 Aug 13;18:116. doi: 10.1186/s12935-018-0602-3. eCollection 2018. Cancer Cell Int. 2018. PMID: 30123094 Free PMC article.
-
Circ_0020123 enhances the cisplatin resistance in non-small cell lung cancer cells partly by sponging miR-140-3p to regulate homeobox B5 (HOXB5).Bioengineered. 2022 Mar;13(3):5126-5140. doi: 10.1080/21655979.2022.2036910. Bioengineered. 2022. PMID: 35170372 Free PMC article.
-
miR-204-5p suppresses hepatocellular cancer proliferation by regulating homeoprotein SIX1 expression.FEBS Open Bio. 2018 Jan 15;8(2):189-200. doi: 10.1002/2211-5463.12363. eCollection 2018 Feb. FEBS Open Bio. 2018. PMID: 29435409 Free PMC article.
-
Transcriptional Repression of CYP3A4 by Increased miR-200a-3p and miR-150-5p Promotes Steatosis in vitro.Front Genet. 2019 May 28;10:484. doi: 10.3389/fgene.2019.00484. eCollection 2019. Front Genet. 2019. PMID: 31191607 Free PMC article.
Cited by
-
Knockdown and Overexpression Experiments to Investigate the Inhibitory Mechanism of Fuzheng Xiaozheng Prescription, an Effective Chinese Herbal Formula for the Clinical Treatment of Hepatocellular Carcinoma.Pharmaceuticals (Basel). 2024 Aug 31;17(9):1159. doi: 10.3390/ph17091159. Pharmaceuticals (Basel). 2024. PMID: 39338323 Free PMC article.
-
miR-9 and miR-181a Target Gab2 to Inhibit the Proliferation and Migration of Hepatocellular Carcinoma HepG2 Cells.Genes (Basel). 2022 Nov 18;13(11):2152. doi: 10.3390/genes13112152. Genes (Basel). 2022. PMID: 36421826 Free PMC article.
-
The homeobox family gene signature predicts the prognosis of osteosarcoma and correlates with immune invasion.Sci Rep. 2025 Jan 6;15(1):886. doi: 10.1038/s41598-024-84924-w. Sci Rep. 2025. PMID: 39762460 Free PMC article.
-
The Circulating miRNA Profile of Chronic Hepatitis D and B Patients Is Comparable but Differs from That of Individuals with HBeAg-Negative HBV Infection.Viruses. 2023 Nov 15;15(11):2257. doi: 10.3390/v15112257. Viruses. 2023. PMID: 38005933 Free PMC article.
-
Uncovering essential anesthetics-induced exosomal miRNAs related to hepatocellular carcinoma progression: a bioinformatic investigation.BMC Med Genomics. 2024 Jun 5;17(1):154. doi: 10.1186/s12920-024-01922-7. BMC Med Genomics. 2024. PMID: 38840234 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

