Essential Dynamics Ensemble Docking for Structure-Based GPCR Drug Discovery
- PMID: 35847975
- PMCID: PMC9277106
- DOI: 10.3389/fmolb.2022.879212
Essential Dynamics Ensemble Docking for Structure-Based GPCR Drug Discovery
Abstract
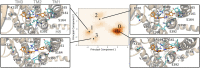
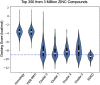
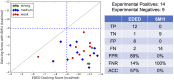
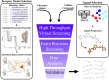
The lack of biologically relevant protein structures can hinder rational design of small molecules to target G protein-coupled receptors (GPCRs). While ensemble docking using multiple models of the protein target is a promising technique for structure-based drug discovery, model clustering and selection still need further investigations to achieve both high accuracy and efficiency. In this work, we have developed an original ensemble docking approach, which identifies the most relevant conformations based on the essential dynamics of the protein pocket. This approach is applied to the study of small-molecule antagonists for the PAC1 receptor, a class B GPCR and a regulator of stress. As few as four representative PAC1 models are selected from simulations of a homology model and then used to screen three million compounds from the ZINC database and 23 experimentally validated compounds for PAC1 targeting. Our essential dynamics ensemble docking (EDED) approach can effectively reduce the number of false negatives in virtual screening and improve the accuracy to seek potent compounds. Given the cost and difficulties to determine membrane protein structures for all the relevant states, our methodology can be useful for future discovery of small molecules to target more other GPCRs, either with or without experimental structures.
Keywords: PAC1 receptor; antagonist; computer aided drug design; molecular dynamics; principal component analysis; virtual screening.
Copyright © 2022 McKay, Hamilton, Remington, Schneebeli and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Improving the Modeling of Extracellular Ligand Binding Pockets in RosettaGPCR for Conformational Selection.Int J Mol Sci. 2023 Apr 24;24(9):7788. doi: 10.3390/ijms24097788. Int J Mol Sci. 2023. PMID: 37175495 Free PMC article.
-
Reliability of Docking-Based Virtual Screening for GPCR Ligands with Homology Modeled Structures: A Case Study of the Angiotensin II Type I Receptor.ACS Chem Neurosci. 2019 Jan 16;10(1):677-689. doi: 10.1021/acschemneuro.8b00489. Epub 2018 Oct 17. ACS Chem Neurosci. 2019. PMID: 30265513
-
Efficiency of Homology Modeling Assisted Molecular Docking in G-protein Coupled Receptors.Curr Top Med Chem. 2021;21(4):269-294. doi: 10.2174/1568026620666200908165250. Curr Top Med Chem. 2021. PMID: 32901584 Review.
-
Novel inhibitor discovery through virtual screening against multiple protein conformations generated via ligand-directed modeling: a maternal embryonic leucine zipper kinase example.J Chem Inf Model. 2012 May 25;52(5):1345-55. doi: 10.1021/ci300040c. Epub 2012 May 8. J Chem Inf Model. 2012. PMID: 22540736
-
Flexible receptor docking for drug discovery.Expert Opin Drug Discov. 2015;10(11):1189-200. doi: 10.1517/17460441.2015.1078308. Epub 2015 Aug 26. Expert Opin Drug Discov. 2015. PMID: 26313123 Review.
Cited by
-
Ensemble Docking as a Tool for the Rational Design of Peptidomimetic Staphylococcus aureus Sortase A Inhibitors.Int J Mol Sci. 2024 Oct 20;25(20):11279. doi: 10.3390/ijms252011279. Int J Mol Sci. 2024. PMID: 39457061 Free PMC article.
-
Revolutionizing Medicinal Chemistry: The Application of Artificial Intelligence (AI) in Early Drug Discovery.Pharmaceuticals (Basel). 2023 Sep 6;16(9):1259. doi: 10.3390/ph16091259. Pharmaceuticals (Basel). 2023. PMID: 37765069 Free PMC article. Review.
-
GPCRLigNet: rapid screening for GPCR active ligands using machine learning.J Comput Aided Mol Des. 2023 Mar;37(3):147-156. doi: 10.1007/s10822-023-00497-2. Epub 2023 Feb 25. J Comput Aided Mol Des. 2023. PMID: 36840893 Free PMC article.
-
Covalent-Allosteric Inhibitors: Do We Get the Best of Both Worlds?J Med Chem. 2025 Feb 27;68(4):4040-4052. doi: 10.1021/acs.jmedchem.4c02760. Epub 2025 Feb 12. J Med Chem. 2025. PMID: 39937154 Free PMC article. Review.
References
-
- Abrol R., Kim S.-K., Bray J. K., Trzaskowski B., Goddard W. A. (2013). “Chapter Two - Conformational Ensemble View of G Protein-Coupled Receptors and the Effect of Mutations and Ligand Binding,” in Methods in Enzymology. Editor Conn P. M. (Academic Press; ), 520, 31–48. 10.1016/b978-0-12-391861-1.00002-2 - DOI - PubMed
-
- Beebe X., Darczak D., Davis-Taber R. A., Uchic M. E., Scott V. E., Jarvis M. F., et al. (2008). Discovery and SAR of Hydrazide Antagonists of the Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP) Receptor Type 1 (PAC1-R). Bioorg. Med. Chem. Lett. 18 (6), 2162–2166. 10.1016/j.bmcl.2008.01.052 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

