Structural Context of a Critical Exon of Spinal Muscular Atrophy Gene
- PMID: 35847983
- PMCID: PMC9283826
- DOI: 10.3389/fmolb.2022.928581
Structural Context of a Critical Exon of Spinal Muscular Atrophy Gene
Abstract
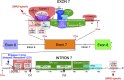
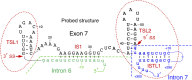
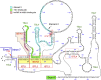
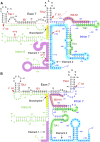
Humans contain two nearly identical copies of Survival Motor Neuron genes, SMN1 and SMN2. Deletion or mutation of SMN1 causes spinal muscular atrophy (SMA), one of the leading genetic diseases associated with infant mortality. SMN2 is unable to compensate for the loss of SMN1 due to predominant exon 7 skipping, leading to the production of a truncated protein. Antisense oligonucleotide and small molecule-based strategies aimed at the restoration of SMN2 exon 7 inclusion are approved therapies of SMA. Many cis-elements and transacting factors have been implicated in regulation of SMN exon 7 splicing. Also, several structural elements, including those formed by a long-distance interaction, have been implicated in the modulation of SMN exon 7 splicing. Several of these structures have been confirmed by enzymatic and chemical structure-probing methods. Additional structures formed by inter-intronic interactions have been predicted by computational algorithms. SMN genes generate a vast repertoire of circular RNAs through inter-intronic secondary structures formed by inverted Alu repeats present in large number in SMN genes. Here, we review the structural context of the exonic and intronic cis-elements that promote or prevent exon 7 recognition. We discuss how structural rearrangements triggered by single nucleotide substitutions could bring drastic changes in SMN2 exon 7 splicing. We also propose potential mechanisms by which inter-intronic structures might impact the splicing outcomes.
Keywords: ISS-N1; RNA structure; SMA; SMN; small molecule; spinal muscular atrophy; splicing; survival motor neuron.
Copyright © 2022 Singh, O'Leary, Eich, Moss and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Evolving concepts on human SMN pre-mRNA splicing.RNA Biol. 2007 Jan-Mar;4(1):7-10. doi: 10.4161/rna.4.1.4535. Epub 2007 Jun 4. RNA Biol. 2007. PMID: 17592254
-
Antisense oligonucleotide mediated therapy of spinal muscular atrophy.Transl Neurosci. 2013 Mar;4(1):10.2478/s13380-013-0109-2. doi: 10.2478/s13380-013-0109-2. Transl Neurosci. 2013. PMID: 24265944 Free PMC article.
-
How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194403. doi: 10.1016/j.bbagrm.2019.07.004. Epub 2019 Jul 16. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31323435 Free PMC article. Review.
-
Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron.Mol Cell Biol. 2006 Feb;26(4):1333-46. doi: 10.1128/MCB.26.4.1333-1346.2006. Mol Cell Biol. 2006. PMID: 16449646 Free PMC article.
-
The First Orally Deliverable Small Molecule for the Treatment of Spinal Muscular Atrophy.Neurosci Insights. 2020 Nov 23;15:2633105520973985. doi: 10.1177/2633105520973985. eCollection 2020. Neurosci Insights. 2020. PMID: 33283185 Free PMC article. Review.
Cited by
-
Ubiquitination Insight from Spinal Muscular Atrophy-From Pathogenesis to Therapy: A Muscle Perspective.Int J Mol Sci. 2024 Aug 13;25(16):8800. doi: 10.3390/ijms25168800. Int J Mol Sci. 2024. PMID: 39201486 Free PMC article. Review.
-
U1 snRNA interactions with deep intronic sequences regulate splicing of multiple exons of spinal muscular atrophy genes.Front Neurosci. 2024 Jul 12;18:1412893. doi: 10.3389/fnins.2024.1412893. eCollection 2024. Front Neurosci. 2024. PMID: 39086841 Free PMC article.
-
How does precursor RNA structure influence RNA processing and gene expression?Biosci Rep. 2023 Mar 31;43(3):BSR20220149. doi: 10.1042/BSR20220149. Biosci Rep. 2023. PMID: 36689327 Free PMC article. Review.
-
Brain-protective mechanisms of autophagy associated circRNAs: Kick starting self-cleaning mode in brain cells via circRNAs as a potential therapeutic approach for neurodegenerative diseases.Front Mol Neurosci. 2023 Jan 16;15:1078441. doi: 10.3389/fnmol.2022.1078441. eCollection 2022. Front Mol Neurosci. 2023. PMID: 36727091 Free PMC article. Review.
-
Discovery of RNA secondary structural motifs using sequence-ordered thermodynamic stability and comparative sequence analysis.MethodsX. 2023 Jun 29;11:102275. doi: 10.1016/j.mex.2023.102275. eCollection 2023 Dec. MethodsX. 2023. PMID: 37448951 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

