Mechanisms of high-level fosfomycin resistance in Staphylococcus aureus epidemic lineage ST5
- PMID: 35848785
- PMCID: PMC9525092
- DOI: 10.1093/jac/dkac236
Mechanisms of high-level fosfomycin resistance in Staphylococcus aureus epidemic lineage ST5
Abstract
Objectives: Fosfomycin resistance has become a clinical concern. In this study, we analysed the dynamic change of fosfomycin MIC in the epidemic Staphylococcus aureus lineages in a teaching hospital in Shanghai for 12 years and sought to elucidate the major underlying mechanisms.
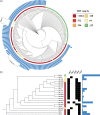
Methods: MLST was conducted for 4580 S. aureus isolates recovered from 2008 to 2019. Fosfomycin MIC was determined by the agar dilution method. The genome data of 230 S. aureus epidemic lineage isolates were acquired from a next-generation sequencing (NGS) platform. Gene deletion and corresponding complementation mutants were constructed to confirm the mechanism of fosfomycin resistance.
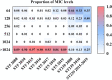
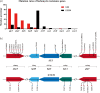
Results: The predominant S. aureus lineages during the past 12 years were ST5 and ST239 (45.6%; 2090/4580). However, ST5 has been spreading clinically, while ST239 has gradually disappeared recently. Consistent with epidemic trends, fosfomycin-resistant ST5 increased from 19.5% to 67.3%. Most fosfomycin-resistant ST5 isolates (92.7%; 647/698) possessed high-level resistance (MIC > 1024 mg/L) with combined mutations mainly in glpT and uhpT. In contrast, fosfomycin-resistant ST239 isolates (76.8%; 149/194) mainly acquired low-level resistance (MIC = 64-128 mg/L) with mutation primarily in hptA. Deletion of a single resistant gene merely resulted in low-level fosfomycin resistance, while double-gene mutants ΔglpTΔuhpT, ΔglpTΔhptA and ΔglpTΔhptR acquired high-level fosfomycin resistance.
Conclusions: The high-level fosfomycin resistance of S. aureus epidemic lineage ST5 is mainly due to the accumulation of mutations in the resistant genes related to membrane transporter systems, and partly contributes to its persistent prevalence under clinical antibiotic pressure.
© The Author(s) 2022. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures


Similar articles
-
hptA Mutation May Mediate Fosfomycin Resistance in Methicillin-Resistant Staphylococcus aureus Clinical Isolates.Microb Drug Resist. 2023 Nov;29(11):497-503. doi: 10.1089/mdr.2022.0173. Epub 2023 Aug 21. Microb Drug Resist. 2023. PMID: 37603296
-
Prevalence of Fosfomycin Resistance and Mutations in murA, glpT, and uhpT in Methicillin-Resistant Staphylococcus aureus Strains Isolated from Blood and Cerebrospinal Fluid Samples.Front Microbiol. 2016 Jan 11;6:1544. doi: 10.3389/fmicb.2015.01544. eCollection 2015. Front Microbiol. 2016. PMID: 26793179 Free PMC article.
-
Prevalence of fosfomycin resistance and gene mutations in clinical isolates of methicillin-resistant Staphylococcus aureus.Antimicrob Resist Infect Control. 2020 Aug 17;9(1):135. doi: 10.1186/s13756-020-00790-x. Antimicrob Resist Infect Control. 2020. PMID: 32807239 Free PMC article.
-
Prevalence of Fosfomycin Resistance in Methicillin-Resistant Staphylococcus aureus Isolated from Patients in a University Hospital in China from 2013 to 2015.Jpn J Infect Dis. 2018 Jul 24;71(4):312-314. doi: 10.7883/yoken.JJID.2018.013. Epub 2018 Apr 27. Jpn J Infect Dis. 2018. PMID: 29709971
-
Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010.J Med Microbiol. 2013 Feb;62(Pt 2):274-282. doi: 10.1099/jmm.0.050971-0. Epub 2012 Nov 1. J Med Microbiol. 2013. PMID: 23118474
Cited by
-
Characteristics of Virulent ST5-SCCmec II Methicillin-Resistant Staphylococcus aureus Prevalent in a Surgery Ward.Infect Drug Resist. 2023 Jun 2;16:3487-3495. doi: 10.2147/IDR.S410330. eCollection 2023. Infect Drug Resist. 2023. PMID: 37293535 Free PMC article.
-
Comparative genomic analysis of antibiotic resistance and virulence genes in Staphylococcus aureus isolates from patients and retail meat.Front Cell Infect Microbiol. 2024 Jan 12;13:1339339. doi: 10.3389/fcimb.2023.1339339. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38282615 Free PMC article.
-
Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus.Antibiotics (Basel). 2022 Oct 8;11(10):1378. doi: 10.3390/antibiotics11101378. Antibiotics (Basel). 2022. PMID: 36290036 Free PMC article. Review.
-
Small regulatory RNA RSaX28 promotes virulence by reinforcing the stability of RNAIII in community-associated ST398 clonotype Staphylococcus aureus.Emerg Microbes Infect. 2024 Dec;13(1):2341972. doi: 10.1080/22221751.2024.2341972. Epub 2024 Apr 20. Emerg Microbes Infect. 2024. PMID: 38597192 Free PMC article.
-
Genetic Complexity of CC5 Staphylococcus aureus Isolates Associated with Sternal Bursitis in Chickens: Antimicrobial Resistance, Virulence, Plasmids, and Biofilm Formation.Pathogens. 2024 Jun 20;13(6):519. doi: 10.3390/pathogens13060519. Pathogens. 2024. PMID: 38921816 Free PMC article.
References
-
- Mehraj J, Witte W, Akmatov MKet al. . Epidemiology of Staphylococcus aureus nasal carriage patterns in the community. Curr Top Microbiol Immunol 2016; 398: 55–87. - PubMed
-
- Wang Y, Lin J, Zhang Tet al. . Environmental contamination prevalence, antimicrobial resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis isolated from secondary schools in Guangzhou, China. Int J Environ Res Public Health 2020; 17: 623. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

