Melanoma RBPome identification reveals PDIA6 as an unconventional RNA-binding protein involved in metastasis
- PMID: 35848924
- PMCID: PMC9371929
- DOI: 10.1093/nar/gkac605
Melanoma RBPome identification reveals PDIA6 as an unconventional RNA-binding protein involved in metastasis
Abstract
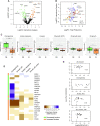
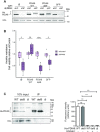
RNA-binding proteins (RBPs) have been relatively overlooked in cancer research despite their contribution to virtually every cancer hallmark. Here, we use RNA interactome capture (RIC) to characterize the melanoma RBPome and uncover novel RBPs involved in melanoma progression. Comparison of RIC profiles of a non-tumoral versus a metastatic cell line revealed prevalent changes in RNA-binding capacities that were not associated with changes in RBP levels. Extensive functional validation of a selected group of 24 RBPs using five different in vitro assays unveiled unanticipated roles of RBPs in melanoma malignancy. As proof-of-principle we focused on PDIA6, an ER-lumen chaperone that displayed a novel RNA-binding activity. We show that PDIA6 is involved in metastatic progression, map its RNA-binding domain, and find that RNA binding is required for PDIA6 tumorigenic properties. These results exemplify how RIC technologies can be harnessed to uncover novel vulnerabilities of cancer cells.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

