Genetic and phenotypic diversity of microsymbionts nodulating promiscuous soybeans from different agro-climatic conditions
- PMID: 35849206
- PMCID: PMC9294079
- DOI: 10.1186/s43141-022-00386-5
Genetic and phenotypic diversity of microsymbionts nodulating promiscuous soybeans from different agro-climatic conditions
Abstract
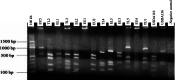
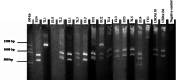
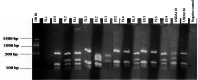
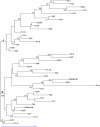
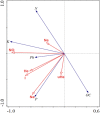
Background: Global food supply is highly dependent on field crop production that is currently severely threatened by changing climate, poor soil quality, abiotic, and biotic stresses. For instance, one of the major challenges to sustainable crop production in most developing countries is limited nitrogen in the soil. Symbiotic nitrogen fixation of legumes such as soybean (Glycine max (L.) Merril) with rhizobia plays a crucial role in supplying nitrogen sufficient to maintain good crop productivity. Characterization of indigenous bradyrhizobia is a prerequisite in the selection and development of effective bioinoculants. In view of this, bradyrhizobia were isolated from soybean nodules in four agro-climatic zones of eastern Kenya (Embu Upper Midland Zone, Embu Lower Midland Zone, Tharaka Upper Midland Zone, and Tharaka Lower Midland Zone) using two soybean varieties (SB8 and SB126). The isolates were characterized using biochemical, morphological, and genotypic approaches. DNA fingerprinting was carried out using 16S rRNA gene and restricted by enzymes HaeIII, Msp1, and EcoRI. RESULTS: Thirty-eight (38) bradyrhizobia isolates obtained from the trapping experiments were placed into nine groups based on their morphological and biochemical characteristics. Most (77%) of the isolates had characteristics of fast-grower bradyrhizobia while 23% were slow-growers. Restriction digest revealed significant (p < 0.015) variation within populations and not among the agro-climatic zones based on analysis of molecular variance. Principal coordinate analysis demonstrated sympatric speciation of indigenous bradyrhizobia isolates. Embu Upper Midland Zone bradyrhizobia isolates had the highest polymorphic loci (80%) and highest genetic diversity estimates (H' = 0.419) compared to other agro-climatic zones.
Conclusion: The high diversity of bradyrhizobia isolates depicts a valuable genetic resource for selecting more effective and competitive strains to improve promiscuous soybean production at a low cost through biological nitrogen fixation.
Keywords: 16S rRNA gene; ARDRA; Bradyrhizobia; Principal coordinate analysis; Shannon’s information index.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no ompeting interests.
Figures


References
-
- Abaidoo RC, Keyser HH, Singleton PW, Dashiell KE, Sanginga N. Population size, distribution, and symbiotic characteristics of indigenous Bradyrhizobium spp. that nodulate TGx soybean genotypes in Africa. Appl Soil Ecol. 2007;35:57–67. doi: 10.1016/j.apsoil.2006.05.006. - DOI
-
- Boddey LH, Hungria M, Hungria M, Megias M. Phenotypic grouping of Brazilian Bradyrhizobium strains which nodulate soybean. Biol Fertil Soils. 1997;25:407–415. doi: 10.1007/s003740050333. - DOI
-
- Bremner JM (1996) Nitrogen‐total. Methods of soil analysis: Part 3 Chemical methods 5:1085–1121
Grants and funding
LinkOut - more resources
Full Text Sources
