Genomic testing for copy number and single nucleotide variants in spermatogenic failure
- PMID: 35849255
- PMCID: PMC9474750
- DOI: 10.1007/s10815-022-02538-5
Genomic testing for copy number and single nucleotide variants in spermatogenic failure
Abstract
Purpose: To identify clinically significant genomic copy number (CNV) and single nucleotide variants (SNV) in males with unexplained spermatogenic failure (SPGF).
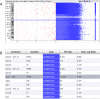
Materials and methods: Peripheral blood DNA from 97/102 study participants diagnosed with oligozoospermia, severe oligozoospermia, or non-obstructive azoospermia (NOA) was analyzed for CNVs via array comparative genomic hybridization (aCGH) and SNVs using whole-exome sequencing (WES).
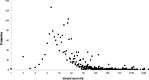
Results: Of the 2544 CNVs identified in individuals with SPGF, > 90% were small, ranging from 0.6 to 75 kb. Thirty, clinically relevant genomic aberrations, were detected in 28 patients (~ 29%). These included likely diagnostic CNVs in 3/41 NOA patients (~ 7%): 1 hemizygous, intragenic TEX11 deletion, 1 hemizygous DDX53 full gene deletion, and 1 homozygous, intragenic STK11 deletion. High-level mosaicism for X chromosome disomy (~ 10% 46,XY and ~ 90% 47,XXY) was also identified in 3 of 41 NOA patients who previously tested normal with conventional karyotyping. The remaining 24 CNVs detected were heterozygous, autosomal recessive carrier variants. Follow-up WES analysis confirmed 8 of 27 (30%) CNVs (X chromosome disomy excluded). WES analysis additionally identified 13 significant SNVs and/or indels in 9 patients (~ 9%) including X-linked AR, KAL1, and NR0B1 variants.
Conclusion: Using a combined genome-wide aCGH/WES approach, we identified pathogenic and likely pathogenic SNVs and CNVs in 15 patients (15%) with unexplained SPGF. This value equals the detection rate of conventional testing for aneuploidies and is considerably higher than the prevalence of Y chromosome microdeletions. Our results underscore the importance of comprehensive genomic analysis in emerging diagnostic testing of complex conditions like male infertility.
Keywords: Comparative genomic hybridization; Copy number variant; Single nucleotide variant; Spermatogenic failure; Whole-exome sequencing.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures
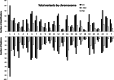


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

