In silico investigation of Alsin RLD conformational dynamics and phosphoinositides binding mechanism
- PMID: 35849605
- PMCID: PMC9292110
- DOI: 10.1371/journal.pone.0270955
In silico investigation of Alsin RLD conformational dynamics and phosphoinositides binding mechanism
Abstract
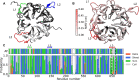
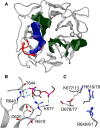
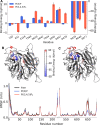
Alsin is a protein known for its major role in neuronal homeostasis and whose mutation is associated with early-onset neurodegenerative diseases. It has been shown that its relocalization from the cytoplasm to the cell membrane is crucial to induce early endosomes maturation. In particular, evidences suggest that the N-terminal regulator of chromosome condensation 1 like domain (RLD) is necessary for membrane association thanks to its affinity to phosphoinositides, membrane lipids involved in the regulation of several signaling processes. Interestingly, this domain showed affinity towards phosphatidylinositol 3-phosphate [PI(3)P], which is highly expressed in endosomes membrane. However, Alsin structure has not been experimentally resolved yet and molecular mechanisms associated with its biological functions are mostly unknown. In this work, Alsin RLD has been investigated through computational molecular modeling techniques to analyze its conformational dynamics and obtain a representative 3D model of this domain. Moreover, a putative phosphoinositide binding site has been proposed and PI(3)P interaction mechanism studied. Results highlight the substantial conformational stability of Alsin RLD secondary structure and suggest the role of one highly flexible region in the phosphoinositides selectivity of this domain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A phosphoinositide conversion mechanism for exit from endosomes.Nature. 2016 Jan 21;529(7586):408-12. doi: 10.1038/nature16516. Epub 2016 Jan 13. Nature. 2016. PMID: 26760201
-
Phosphoinositides differentially regulate protrudin localization through the FYVE domain.J Biol Chem. 2012 Nov 30;287(49):41268-76. doi: 10.1074/jbc.M112.419127. Epub 2012 Oct 5. J Biol Chem. 2012. PMID: 23043110 Free PMC article.
-
Phosphoinositides and membrane traffic at the trans-Golgi network.Biochem Soc Symp. 2005;(72):31-8. doi: 10.1042/bss0720031. Biochem Soc Symp. 2005. PMID: 15649127 Review.
-
Receptor-mediated Endocytosis 8 Utilizes an N-terminal Phosphoinositide-binding Motif to Regulate Endosomal Clathrin Dynamics.J Biol Chem. 2015 Aug 28;290(35):21676-89. doi: 10.1074/jbc.M115.644757. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134565 Free PMC article.
-
Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways.Int J Mol Sci. 2017 Mar 15;18(3):634. doi: 10.3390/ijms18030634. Int J Mol Sci. 2017. PMID: 28294977 Free PMC article. Review.
Cited by
-
VirtuousPocketome: a computational tool for screening protein-ligand complexes to identify similar binding sites.Sci Rep. 2024 Mar 15;14(1):6296. doi: 10.1038/s41598-024-56893-7. Sci Rep. 2024. PMID: 38491261 Free PMC article.
-
Spastin and alsin protein interactome analyses begin to reveal key canonical pathways and suggest novel druggable targets.Neural Regen Res. 2025 Mar 1;20(3):725-739. doi: 10.4103/NRR.NRR-D-23-02068. Epub 2024 May 13. Neural Regen Res. 2025. PMID: 38886938 Free PMC article.
References
-
- Sato K, Otomo A, Ueda MT, Hiratsuka Y, Suzuki-Utsunomiya K, Sugiyama J, et al.. Altered oligomeric states in pathogenic ALS2 variants associated with juvenile motor neuron diseases cause loss of ALS2-mediated endosomal function. J Biol Chem. 2018;293: 17135–17153. doi: 10.1074/jbc.RA118.003849 - DOI - PMC - PubMed
-
- Orrell RW. ALS2-Related Disorder. GeneReviews®. 1993. Available: http://www.ncbi.nlm.nih.gov/pubmed/20301421 - PubMed
-
- Otomo A, Kunita R, Suzuki-Utsunomiya K, Ikeda J-E, Hadano S. Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett. 2011;585: 730–6. doi: 10.1016/j.febslet.2011.01.045 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

