Design, Synthesis, and Biological Activity of New CB2 Receptor Ligands: from Orthosteric and Allosteric Modulators to Dualsteric/Bitopic Ligands
- PMID: 35849804
- PMCID: PMC10168668
- DOI: 10.1021/acs.jmedchem.2c00582
Design, Synthesis, and Biological Activity of New CB2 Receptor Ligands: from Orthosteric and Allosteric Modulators to Dualsteric/Bitopic Ligands
Abstract
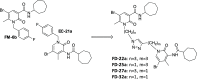
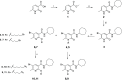
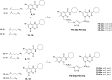
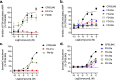
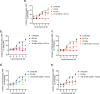
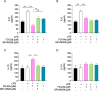
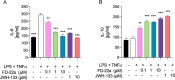
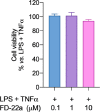
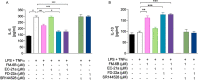
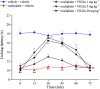
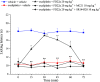
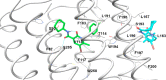
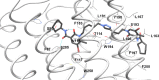
The design of dualsteric/bitopic agents as single chemical entities able to simultaneously interact with both the orthosteric and an allosteric binding site represents a novel approach in medicinal chemistry. Biased dualsteric/bitopic agents could enhance certain signaling pathways while diminishing the others that cause unwanted side effects. We have designed, synthesized, and functionally characterized the first CB2R heterobivalent bitopic ligands. In contrast to the parent orthosteric compound, our bitopic ligands selectively target CB2R versus CB1R and show a functional selectivity for the cAMP signaling pathway versus βarrestin2 recruitment. Moreover, the most promising bitopic ligand FD-22a displayed anti-inflammatory activity in a human microglial cell inflammatory model and antinociceptive activity in vivo in an experimental mouse model of neuropathic pain. Finally, computational studies clarified the binding mode of these compounds inside the CB2R, further confirming their bitopic nature.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
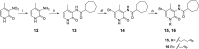

Similar articles
-
New Insights into Bitopic Orthosteric/Allosteric Ligands of Cannabinoid Receptor Type 2.Int J Mol Sci. 2023 Jan 21;24(3):2135. doi: 10.3390/ijms24032135. Int J Mol Sci. 2023. PMID: 36768458 Free PMC article.
-
Design, synthesis and biological evaluation of novel orthosteric-allosteric ligands of the cannabinoid receptor type 2 (CB2R).Front Chem. 2022 Sep 27;10:984069. doi: 10.3389/fchem.2022.984069. eCollection 2022. Front Chem. 2022. PMID: 36238097 Free PMC article.
-
Bridging the Binding Sites 2.0: Photoswitchable Dualsteric Ligands for the Cannabinoid 2 Receptor.ACS Chem Neurosci. 2023 Oct 18;14(20):3737-3744. doi: 10.1021/acschemneuro.3c00509. Epub 2023 Oct 4. ACS Chem Neurosci. 2023. PMID: 37792463
-
Molecular alliance-from orthosteric and allosteric ligands to dualsteric/bitopic agonists at G protein coupled receptors.Angew Chem Int Ed Engl. 2013 Jan 7;52(2):508-16. doi: 10.1002/anie.201205315. Epub 2012 Dec 6. Angew Chem Int Ed Engl. 2013. PMID: 23225228 Review.
-
Computational Advances for the Development of Allosteric Modulators and Bitopic Ligands in G Protein-Coupled Receptors.AAPS J. 2015 Sep;17(5):1080-95. doi: 10.1208/s12248-015-9776-y. Epub 2015 May 5. AAPS J. 2015. PMID: 25940084 Free PMC article. Review.
Cited by
-
The Skin and Natural Cannabinoids-Topical and Transdermal Applications.Pharmaceuticals (Basel). 2023 Jul 24;16(7):1049. doi: 10.3390/ph16071049. Pharmaceuticals (Basel). 2023. PMID: 37513960 Free PMC article. Review.
-
Novel Cannabinoid Receptor 2 (CB2) Low Lipophilicity Agonists Produce Distinct cAMP and Arrestin Signalling Kinetics without Bias.Int J Mol Sci. 2023 Mar 29;24(7):6406. doi: 10.3390/ijms24076406. Int J Mol Sci. 2023. PMID: 37047385 Free PMC article.
-
Development and evaluation of deuterated [18F]JHU94620 isotopologues for the non-invasive assessment of the cannabinoid type 2 receptor in brain.EJNMMI Radiopharm Chem. 2024 Dec 23;9(1):91. doi: 10.1186/s41181-024-00319-2. EJNMMI Radiopharm Chem. 2024. PMID: 39714717 Free PMC article.
-
T1AM/TAAR1 System Reduces Inflammatory Response and β-Amyloid Toxicity in Human Microglial HMC3 Cell Line.Int J Mol Sci. 2023 Jul 17;24(14):11569. doi: 10.3390/ijms241411569. Int J Mol Sci. 2023. PMID: 37511328 Free PMC article.
-
A Proteomic Approach Identified TFEB as a Key Player in the Protective Action of Novel CB2R Bitopic Ligand FD22a against the Deleterious Effects Induced by β-Amyloid in Glial Cells.Cells. 2024 May 19;13(10):875. doi: 10.3390/cells13100875. Cells. 2024. PMID: 38786097 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information

