An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients With Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study
- PMID: 35850192
- PMCID: PMC9613527
- DOI: 10.1053/j.gastro.2022.06.090
An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients With Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study
Abstract
Background & aims: Pancreatic ductal adenocarcinoma (PDAC) incidence is rising worldwide, and most patients present with an unresectable disease at initial diagnosis. Measurement of carbohydrate antigen 19-9 (CA19-9) levels lacks adequate sensitivity and specificity for early detection; hence, there is an unmet need to develop alternate molecular diagnostic biomarkers for PDAC. Emerging evidence suggests that tumor-derived exosomal cargo, particularly micro RNAs (miRNAs), offer an attractive platform for the development of cancer-specific biomarkers. Herein, genomewide profiling in blood specimens was performed to develop an exosome-based transcriptomic signature for noninvasive and early detection of PDAC.
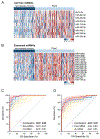
Methods: Small RNA sequencing was undertaken in a cohort of 44 patients with an early-stage PDAC and 57 nondisease controls. Using machine-learning algorithms, a panel of cell-free (cf) and exosomal (exo) miRNAs were prioritized that discriminated patients with PDAC from control subjects. Subsequently, the performance of the biomarkers was trained and validated in independent cohorts (n = 191) using quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays.
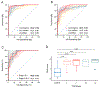
Results: The sequencing analysis initially identified a panel of 30 overexpressed miRNAs in PDAC. Subsequently using qRT-PCR assays, the panel was reduced to 13 markers (5 cf- and 8 exo-miRNAs), which successfully identified patients with all stages of PDAC (area under the curve [AUC] = 0.98 training cohort; AUC = 0.93 validation cohort); but more importantly, was equally robust for the identification of early-stage PDAC (stages I and II; AUC = 0.93). Furthermore, this transcriptomic signature successfully identified CA19-9 negative cases (<37 U/mL; AUC = 0.96), when analyzed in combination with CA19-9 levels, significantly improved the overall diagnostic accuracy (AUC = 0.99 vs AUC = 0.86 for CA19-9 alone).
Conclusions: In this study, an exosome-based liquid biopsy signature for the noninvasive and robust detection of patients with PDAC was developed.
Keywords: Diagnostic Biomarker; Exosome; Liquid Biopsy; Pancreatic Cancer; miRNA Signature.
Copyright © 2022 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest: None of the authors has any potential conflicts to disclose.
Competing interests: The authors declare that they have no competing interests.
Figures


Comment in
-
Early Detection of Pancreatic Cancer: Are We Ready for Prime Time?Gastroenterology. 2022 Nov;163(5):1157-1159. doi: 10.1053/j.gastro.2022.07.072. Epub 2022 Aug 2. Gastroenterology. 2022. PMID: 35931105 No abstract available.
References
-
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039–49. - PubMed
-
- Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022. - PubMed
-
- Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. The Lancet 2020;395:2008–2020. - PubMed
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–249. - PubMed
-
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

