Wastewater surveillance allows early detection of SARS-CoV-2 omicron in North Rhine-Westphalia, Germany
- PMID: 35850355
- PMCID: PMC9287496
- DOI: 10.1016/j.scitotenv.2022.157375
Wastewater surveillance allows early detection of SARS-CoV-2 omicron in North Rhine-Westphalia, Germany
Abstract
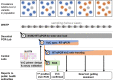
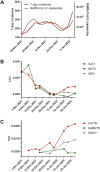
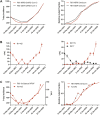
Wastewater-based epidemiology (WBE) has demonstrated its importance to support SARS-CoV-2 epidemiology complementing individual testing strategies. Due to their immune-evasive potential and the resulting significance for public health, close monitoring of SARS-CoV-2 variants of concern (VoC) is required to evaluate the regulation of early local countermeasures. In this study, we demonstrate a rapid workflow for wastewater-based early detection and monitoring of the newly emerging SARS-CoV-2 VoCs Omicron in the end of 2021 at the municipal wastewater treatment plant (WWTP) Emschermuendung (KLEM) in the Federal State of North-Rhine-Westphalia (NRW, Germany). Initially, available primers detecting Omicron-related mutations were rapidly validated in a central laboratory. Subsequently, RT-qPCR analysis of purified SARS-CoV-2 RNA was performed in a decentral PCR laboratory in close proximity to KLEM. This decentralized approach enabled the early detection of K417N present in Omicron in samples collected on 8th December 2021 and the detection of further mutations (N501Y, Δ69/70) in subsequent biweekly sampling campaigns. The presence of Omicron in wastewater was confirmed by next generation sequencing (NGS) in a central laboratory with samples obtained on 14th December 2021. Moreover, the relative increase of the mutant fraction of Omicron was quantitatively monitored over time by dPCR in a central PCR laboratory starting on 12th December 2021 confirming Omicron as the dominant variant by the end of 2021. In conclusions, WBE plays a crucial role in surveillance of SARS-CoV-2 variants and is suitable as an early warning system to identify variant emergence. In particular, the successive workflow using RT-qPCR, RT-dPCR and NGS demonstrates the strength of WBE as a versatile tool to monitor variant spreading.
Keywords: COVID-19 surveillance; Detection workflow; Omicron, B.1.1.529; SARS-CoV-2 monitoring; Variant of concern; Wastewater-based epidemiology (WBE).
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Alexander Wilhelm reports equipment, drugs, or supplies were provided by QIAGEN GmbH. Qiagen GmbH and Endress+Hauser are associated industry partner of the COVIDready consortium. Jens Schoth and Burkhardt Teichgräber declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Qiagen GmbH and Endress+Hauser are associated industry partner of the COVIDready consortium. Marek Widera reports equipment, drugs, or supplies were provided by QIAGEN GmbH. Qiagen GmbH and Endress+Hauser are associated industry partner of the COVIDready consortium.
Figures

References
-
- Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 omicron variant at Frankfurt airport but limited spread in the City of Frankfurt, Germany, in november 2021. Microbiol. Resour. Announc. 2022;11 - PMC - PubMed
-
- Bonanno Ferraro G., Veneri C., Mancini P., Iaconelli M., Suffredini E., Bonadonna L., et al. A state-of-the-art scoping review on SARS-CoV-2 in sewage focusing on the potential of wastewater surveillance for the monitoring of the COVID-19 pandemic. Food Environ. Virol. 2021:1–40. doi: 10.1007/s12560-021-09498-6. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

