Combinatorial targeting of a chromatin complex comprising Dot1L, menin and the tyrosine kinase BAZ1B reveals a new therapeutic vulnerability of endocrine therapy-resistant breast cancer
- PMID: 35850772
- PMCID: PMC9290241
- DOI: 10.1186/s13058-022-01547-7
Combinatorial targeting of a chromatin complex comprising Dot1L, menin and the tyrosine kinase BAZ1B reveals a new therapeutic vulnerability of endocrine therapy-resistant breast cancer
Abstract
Background: Targeting vulnerabilities of cancer cells by inhibiting key regulators of cell proliferation or survival represents a promising way to overcome resistance to current therapies. In breast cancer (BC), resistance to endocrine therapy results from constitutively active or aberrant estrogen receptor alpha (ERα) signaling to the genome. Targeting components of the ERα pathway in these tumors represents, therefore, a rational way toward effective new treatments. Interaction proteomics identified several proteins associated with ERα in BC cells, including epigenetic complexes controlling gene transcription comprising the scaffold protein menin and the histone methyltransferase Dot1L.
Methods: We combined chromatin immunoprecipitation, transcriptome sequencing, siRNA-mediated gene knockdown (kd), pharmacological inhibition coupled to cellular and functional assays and interaction proteomics in antiestrogen (AE)-sensitive and AE-resistant human BC cell models to: map menin and Dot1L chromatin localization, search for their common and specific target genes, measure the effects of single or combinatorial knockdown or pharmacological inhibition of these proteins on cell proliferation and survival, and characterize their nuclear interactomes.
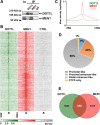
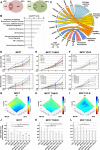
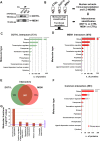
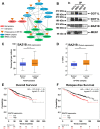
Results: Dot1L and menin associate in MCF-7 cells chromatin, where they co-localize in a significant fraction of sites, resulting in co-regulation of genes involved, among others, in estrogen, p53, HIF1α and death receptor signaling, regulation of cell cycle and epithelial-to-mesenchymal transition. Specific inhibitors of the two factors synergize with each other for inhibition of cell proliferation of AE (tamoxifen or fulvestrant)-sensitive and AE-resistant BC cells. Menin and Dot1L interactomes share a sizeable fraction of their nuclear partners, the majority being known BC fitness genes. Interestingly, these include B-WICH and WINAC complexes that share BAZ1B, a bromodomain protein comprising a tyrosine-protein kinase domain playing a central role in chromatin remodeling and transcriptional regulation. BAZ1B kd caused significant inhibition of ERα expression, proliferation and transcriptome changes resulting in inhibition of estrogen, myc, mTOR, PI3K and AKT signaling and metabolic pathways in AE-sensitive and AE-resistant BC cells.
Conclusions: Identification of a functional interplay between ERα, Dot1L, menin and BAZ1B and the significant effects of their co-inhibition on cell proliferation and survival in cell models of endocrine therapy-resistant BC reveal a new therapeutic vulnerability of these aggressive diseases.
Keywords: BAZ1B; Breast cancer; Dot1L; Endocrine therapy resistance; Estrogen signaling; Menin.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

