Reconstruction of transmission chains of SARS-CoV-2 amidst multiple outbreaks in a geriatric acute-care hospital: a combined retrospective epidemiological and genomic study
- PMID: 35850933
- PMCID: PMC9328768
- DOI: 10.7554/eLife.76854
Reconstruction of transmission chains of SARS-CoV-2 amidst multiple outbreaks in a geriatric acute-care hospital: a combined retrospective epidemiological and genomic study
Abstract
Background: There is ongoing uncertainty regarding transmission chains and the respective roles of healthcare workers (HCWs) and elderly patients in nosocomial outbreaks of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in geriatric settings.
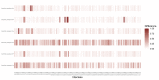
Methods: We performed a retrospective cohort study including patients with nosocomial coronavirus disease 2019 (COVID-19) in four outbreak-affected wards, and all SARS-CoV-2 RT-PCR positive HCWs from a Swiss university-affiliated geriatric acute-care hospital that admitted both Covid-19 and non-Covid-19 patients during the first pandemic wave in Spring 2020. We combined epidemiological and genetic sequencing data using a Bayesian modelling framework, and reconstructed transmission dynamics of SARS-CoV-2 involving patients and HCWs, to determine who infected whom. We evaluated general transmission patterns according to case type (HCWs working in dedicated Covid-19 cohorting wards: HCWcovid; HCWs working in non-Covid-19 wards where outbreaks occurred: HCWoutbreak; patients with nosocomial Covid-19: patientnoso) by deriving the proportion of infections attributed to each case type across all posterior trees and comparing them to random expectations.
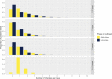
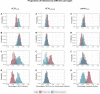
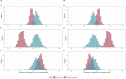
Results: During the study period (1 March to 7 May 2020), we included 180 SARS-CoV-2 positive cases: 127 HCWs (91 HCWcovid, 36 HCWoutbreak) and 53 patients. The attack rates ranged from 10% to 19% for patients, and 21% for HCWs. We estimated that 16 importation events occurred with high confidence (4 patients, 12 HCWs) that jointly led to up to 41 secondary cases; in six additional cases (5 HCWs, 1 patient), importation was possible with a posterior probability between 10% and 50%. Most patient-to-patient transmission events involved patients having shared a ward (95.2%, 95% credible interval [CrI] 84.2%-100%), in contrast to those having shared a room (19.7%, 95% CrI 6.7%-33.3%). Transmission events tended to cluster by case type: patientnoso were almost twice as likely to be infected by other patientnoso than expected (observed:expected ratio 2.16, 95% CrI 1.17-4.20, p=0.006); similarly, HCWoutbreak were more than twice as likely to be infected by other HCWoutbreak than expected (2.72, 95% CrI 0.87-9.00, p=0.06). The proportion of infectors being HCWcovid was as expected as random. We found a trend towards a greater proportion of high transmitters (≥2 secondary cases) among HCWoutbreak than patientnoso in the late phases (28.6% vs. 11.8%) of the outbreak, although this was not statistically significant.
Conclusions: Most importation events were linked to HCW. Unexpectedly, transmission between HCWcovid was more limited than transmission between patients and HCWoutbreak. This finding highlights gaps in infection control and suggests the possible areas of improvements to limit the extent of nosocomial transmission.
Funding: This study was supported by a grant from the Swiss National Science Foundation under the NRP78 funding scheme (Grant no. 4078P0_198363).
Keywords: COVID-19; SARS-CoV-2; geriatric hospitals; healthcare-associated infection; infection prevention; infectious disease; medicine; microbiology; nosocomial outbreaks; transmission dynamics; viruses.
© 2022, Abbas et al.
Conflict of interest statement
MA, SC, FL, TR, AM, JS, PH, DZ, VP, AI, LV, VS, CG, SH No competing interests declared, AC received honoraria (which was paid to the institution) from Pfizer for lecturing on a course on mathematical modelling of infectious disease transmission and vaccination book. The author has no other competing interests to declare
Figures








References
-
- Abbas M, Robalo Nunes T, Cori A, Cordey S, Laubscher F, Baggio S, Jombart T, Iten A, Vieux L, Teixeira D, Perez M, Pittet D, Frangos E, Graf CE, Zingg W, Harbarth S. Explosive nosocomial outbreak of SARS-CoV-2 in a rehabilitation clinic: the limits of genomics for outbreak reconstruction. The Journal of Hospital Infection. 2021a;117:124–134. doi: 10.1016/j.jhin.2021.07.013. - DOI - PMC - PubMed
-
- Abbas M, Robalo Nunes T, Martischang R, Zingg W, Iten A, Pittet D, Harbarth S. Nosocomial transmission and outbreaks of coronavirus disease 2019: the need to protect both patients and healthcare workers. Antimicrobial Resistance and Infection Control. 2021b;10:7. doi: 10.1186/s13756-020-00875-7. - DOI - PMC - PubMed
-
- Aggarwal D, Myers R, Hamilton WL, Bharucha T, Tumelty NM, Brown CS, Meader EJ, Connor T, Smith DL, Bradley DT, Robson S, Bashton M, Shallcross L, Zambon M, Goodfellow I, Chand M, O’Grady J, Török ME, Peacock SJ, Page AJ, COVID-19 Genomics UK (COG-UK) Consortium The role of viral genomics in understanding COVID-19 outbreaks in long-term care facilities. The Lancet. Microbe. 2022;3:e151–e158. doi: 10.1016/S2666-5247(21)00208-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

