PARP14 is a novel target in STAT6 mutant follicular lymphoma
- PMID: 35851155
- PMCID: PMC9417990
- DOI: 10.1038/s41375-022-01641-x
PARP14 is a novel target in STAT6 mutant follicular lymphoma
Abstract
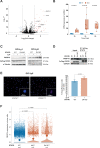
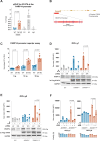
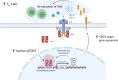
The variable clinical course of follicular lymphoma (FL) is determined by the molecular heterogeneity of tumor cells and complex interactions within the tumor microenvironment (TME). IL-4 producing follicular helper T cells (TFH) are critical components of the FL TME. Binding of IL-4 to IL-4R on FL cells activates JAK/STAT signaling. We identified STAT6 mutations (STAT6MUT) in 13% of FL (N = 33/258), all clustered within the DNA binding domain. Gene expression data and immunohistochemistry showed upregulation of IL-4/STAT6 target genes in STAT6MUT FL, including CCL17, CCL22, and FCER2 (CD23). Functionally, STAT6MUT was gain-of-function by serial replating phenotype in pre-B CFU assays. Expression of STAT6MUT enhanced IL-4 induced FCER2/CD23, CCL17 and CCL22 expression and was associated with nuclear accumulation of pSTAT6. RNA sequencing identified PARP14 -a transcriptional switch and co-activator of STAT6- among the top differentially upregulated genes in IL-4 stimulated STAT6MUT lymphoma cells and in STAT6MUT primary FL cells. Quantitative chromatin immunoprecipitation (qChIP) demonstrated binding of STAT6MUT but not STAT6WT to the PARP14 promotor. Reporter assays showed increased IL-4 induced transactivation activity of STAT6MUT at the PARP14 promotor, suggesting a self-reinforcing regulatory circuit. Knock-down of PARP14 or PARP-inhibition abrogated the STAT6MUT gain-of-function phenotype. Thus, our results identify PARP14 as a novel therapeutic target in STAT6MUT FL.
© 2022. The Author(s).
Conflict of interest statement
This research was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Project-ID WE 4679/2-1 (OW), SCHM2440/8-1 (MSS), and the Max-Eder Program of the Deutsche Krebshilfe e.V. (DKH, German Cancer Aid), Project-ID 110659/70112997 (OW). OW is supported by an Else-Kröner Excellence Fellowship from the Else-Kröner-Fresenius Stiftung (Project-ID 2021_EKES.13). LA is supported by the Else Kröner-Fresenius Clinician Scientist Program: Cancer immunotherapy, and the Munich Clinician Scientist Program (MCSP). LA and JAH are supported by the German Consortium for Translational Cancer Research (DKTK) School of Oncology. MS, Amgen, Speakers Bureau. The authors declare no competing interests.
Figures



References
-
- Casulo C, Byrtek M, Dawson KL, Zhou X, Farber CM, Flowers CR, et al. Early Relapse of Follicular Lymphoma After Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone Defines Patients at High Risk for Death: An Analysis From the National LymphoCare Study. J Clin Oncol : Off J Am Soc Clin Oncol. 2015;33:2516–22. doi: 10.1200/JCO.2014.59.7534. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

