A strategy to quantify myofibroblast activation on a continuous spectrum
- PMID: 35851602
- PMCID: PMC9293987
- DOI: 10.1038/s41598-022-16158-7
A strategy to quantify myofibroblast activation on a continuous spectrum
Abstract
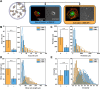
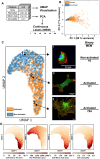
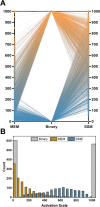
Myofibroblasts are a highly secretory and contractile cell phenotype that are predominant in wound healing and fibrotic disease. Traditionally, myofibroblasts are identified by the de novo expression and assembly of alpha-smooth muscle actin stress fibers, leading to a binary classification: "activated" or "quiescent (non-activated)". More recently, however, myofibroblast activation has been considered on a continuous spectrum, but there is no established method to quantify the position of a cell on this spectrum. To this end, we developed a strategy based on microscopy imaging and machine learning methods to quantify myofibroblast activation in vitro on a continuous scale. We first measured morphological features of over 1000 individual cardiac fibroblasts and found that these features provide sufficient information to predict activation state. We next used dimensionality reduction techniques and self-supervised machine learning to create a continuous scale of activation based on features extracted from microscopy images. Lastly, we compared our findings for mechanically activated cardiac fibroblasts to a distribution of cell phenotypes generated from transcriptomic data using single-cell RNA sequencing. Altogether, these results demonstrate a continuous spectrum of myofibroblast activation and provide an imaging-based strategy to quantify the position of a cell on that spectrum.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

