Ability of Two Strains of Lactic Acid Bacteria To Inhibit Listeria monocytogenes by Spot Inoculation and in an Environmental Microbiome Context
- PMID: 35852346
- PMCID: PMC9431016
- DOI: 10.1128/spectrum.01018-22
Ability of Two Strains of Lactic Acid Bacteria To Inhibit Listeria monocytogenes by Spot Inoculation and in an Environmental Microbiome Context
Abstract
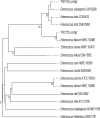
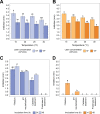
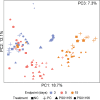
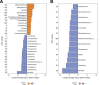
We evaluated the ability of two strains of lactic acid bacteria (LAB) to inhibit L. monocytogenes using spot inoculation and environmental microbiome attached-biomass assays. LAB strains (PS01155 and PS01156) were tested for antilisterial activity toward 22 phylogenetically distinct L. monocytogenes strains isolated from three fruit packing environments (F1, F2, and F3). LAB strains were tested by spot inoculation onto L. monocytogenes lawns (108 and 107 CFU/mL) and incubated at 15, 20, 25, or 30°C for 3 days. The same LAB strains were also cocultured at 15°C for 3, 5, and 15 days in polypropylene conical tubes with L. monocytogenes and environmental microbiome suspensions collected from F1, F2, and F3. In the spot inoculation assay, PS01156 was significantly more inhibitory toward less concentrated L. monocytogenes lawns than more concentrated lawns at all the tested temperatures, while PS01155 was significantly more inhibitory toward less concentrated lawns only at 15 and 25°C. Furthermore, inhibition of L. monocytogenes by PS01156 was significantly greater at 15°C than higher temperatures, whereas the temperature did not have an effect on the inhibitory activity of PS01155. In the assay using attached environmental microbiome biomass, L. monocytogenes concentration was significantly reduced by PS01156, but not PS01155, when cocultured with microbiomes from F1 and F3 and incubated for 3 days at 15°C. Attached biomass microbiota composition was significantly affected by incubation time but not by LAB strain. This study demonstrates that LAB strains that may exhibit inhibitory properties toward L. monocytogenes in a spot inoculation assay may not maintain antilisterial activity within a complex microbiome. IMPORTANCE Listeria monocytogenes has previously been associated with outbreaks of foodborne illness linked to consumption of fresh produce. In addition to conventional cleaning and sanitizing, lactic acid bacteria (LAB) have been studied for biocontrol of L. monocytogenes in food processing environments that are challenging to clean and sanitize. We evaluated whether two specific LAB strains, PS01155 and PS01156, can inhibit the growth of L. monocytogenes strains in a spot inoculation and in an attached-biomass assay, in which they were cocultured with environmental microbiomes collected from tree fruit packing facilities. LAB strains PS01155 and PS01156 inhibited L. monocytogenes in a spot inoculation assay, but the antilisterial activity was lower or not detected when they were grown with environmental microbiota. These results highlight the importance of conducting biocontrol challenge tests in the context of the complex environmental microbiomes present in food processing facilities to assess their potential for application in the food industry.
Keywords: Listeria monocytogenes; attached biomass; biocontrol; biofilms; environmental microbiome; inhibition; lactic acid bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- CDC. 2018. Burden of foodborne illness: findings. https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html. Retrieved 4 June 2020.
-
- Mazaheri T, Cervantes-Huamán BRH, Bermúdez-Capdevila M, Ripolles-Avila C, Rodríguez-Jerez JJ. 2021. Listeria monocytogenes biofilms in the food industry: is the current hygiene program sufficient to combat the persistence of the pathogen? Microorganisms 9:181. doi: 10.3390/microorganisms9010181. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

