First Genotype-Phenotype Study in TBX4 Syndrome: Gain-of-Function Mutations Causative for Lung Disease
- PMID: 35852389
- PMCID: PMC9757087
- DOI: 10.1164/rccm.202203-0485OC
First Genotype-Phenotype Study in TBX4 Syndrome: Gain-of-Function Mutations Causative for Lung Disease
Abstract
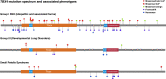
Rationale: Despite the increased recognition of TBX4 (T-BOX transcription factor 4)-associated pulmonary arterial hypertension (PAH), genotype-phenotype associations are lacking and may provide important insights. Objectives: To compile and functionally characterize all TBX4 variants reported to date and undertake a comprehensive genotype-phenotype analysis. Methods: We assembled a multicenter cohort of 137 patients harboring monoallelic TBX4 variants and assessed the pathogenicity of missense variation (n = 42) using a novel luciferase reporter assay containing T-BOX binding motifs. We sought genotype-phenotype correlations and undertook a comparative analysis with patients with PAH with BMPR2 (Bone Morphogenetic Protein Receptor type 2) causal variants (n = 162) or no identified variants in PAH-associated genes (n = 741) genotyped via the National Institute for Health Research BioResource-Rare Diseases. Measurements and Main Results: Functional assessment of TBX4 missense variants led to the novel finding of gain-of-function effects associated with older age at diagnosis of lung disease compared with loss-of-function effects (P = 0.038). Variants located in the T-BOX and nuclear localization domains were associated with earlier presentation (P = 0.005) and increased incidence of interstitial lung disease (P = 0.003). Event-free survival (death or transplantation) was shorter in the T-BOX group (P = 0.022), although age had a significant effect in the hazard model (P = 0.0461). Carriers of TBX4 variants were diagnosed at a younger age (P < 0.001) and had worse baseline lung function (FEV1, FVC) (P = 0.009) than the BMPR2 and no identified causal variant groups. Conclusions: We demonstrated that TBX4 syndrome is not strictly the result of haploinsufficiency but can also be caused by gain of function. The pleiotropic effects of TBX4 in lung disease may be in part explained by the differential effect of pathogenic mutations located in critical protein domains.
Keywords: TBX4; gain-of-function; interstitial lung disease; lung developmental disease; pulmonary arterial hypertension.
Figures




Comment in
-
Understanding Genotype-Phenotype Correlations in Patients with TBX4 Mutations: New Views Inside and Outside the Box.Am J Respir Crit Care Med. 2022 Dec 15;206(12):1448-1449. doi: 10.1164/rccm.202208-1461ED. Am J Respir Crit Care Med. 2022. PMID: 35925028 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

